Abstract
Plasticity of root growth in response to environmental cues and stresses is a fundamental characteristic of plants, in accordance with their sessile lifestyle. This is linked to the balance between plasticity and rigidity of cells in the root apex, and thus is coordinated with the control of cell wall properties. However, mechanisms underlying such harmonization are not well understood, in particular under stressful conditions. We have recently demonstrated that RICE SALT SENSITIVE3 (RSS3), a nuclear factor that mediates restrictive expression of jasmonate-induced genes, plays an important role in root elongation under saline conditions. In this study, we report that loss-of-function of RSS3 results in changes in cell wall properties such as lignin deposition and sensitivity to a cellulose synthase inhibitor, concomitant with altered expression of genes involved in cell wall metabolism. Based on these and previous phenotypic observations of the rss3 mutant, we propose that RSS3 plays a role in the coordinated control of root elongation and cell wall plasticity in the root apex.
Growth and development of plant roots is sustained by continuous cell division at the root apex and subsequent cell differentiation. The direction and extent of root elongation is flexible and responsive to various environmental factors such as gravity, water and nutrient availability, and stressful conditions. To ensure the flexibility of root growth, it is important for the plant to maintain a balance between rigidity and plasticity of root cells, which may be correlated with cell wall properties. Rigidity or plasticity of cell wall may affect not only cell division and cell elongation but also the organization of differentiated cells. Cell walls are synthesized or reconstituted, depending on the process that cells undergo, whereas physical and chemical structures of cell walls are altered by environmental conditions.Citation1,Citation2 For example, in tobacco cells, the cell wall exhibits weakened tensile strength under high-salinity conditions, which is associated with a decrease in cellulose content, compositional changes in cell wall-associated proteins, and reorganization of pectin.Citation1 However, it is largely unknown how the coordinated control between growth of root cells and cell wall properties is achieved.
RICE SALT SENSITIVE3 (RSS3) is a nuclear factor that interacts and forms a ternary complex with class-C bHLH transcription factors and JASMONATE ZIM DOMAIN (JAZ) proteins.Citation3 A loss-of-function mutant of RSS3 (rss3) was originally identified in a screen of salt-sensitive mutants.Citation4 Under conditions without salinity stress, rss3 shows a moderate reduction in root tip cell elongation but exhibits severe inhibition of root growth under salinity stressed conditions.Citation3 This is accompanied by aberrant cellular arrangement, formation of oblique cell plates, swelling of cells, and impaired root flexibility. Cell swelling and abnormal cytokinesis are also observed in cellulose-deficient mutants such as radial swelling1 (rsw1) and rsw2.Citation5,Citation6 In addition, reduced root cell elongation and aberrant cell expansion are observed in the cobra mutant deficient in cellulose microfibril organizationCitation7 or roots grown in the presence of isoxaben, a cellulose synthase inhibitor.Citation8 These observations raise the possibility that the rss3 mutant has a defect in the cell wall.
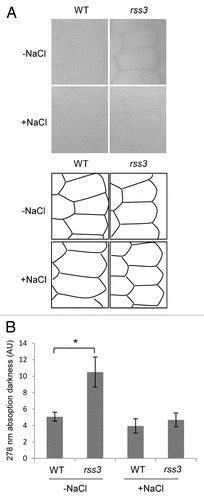
In the previous study, it was revealed by transcriptomic analysis of the root tip and subsequent enrichment analysis that many genes involved in cell wall metabolism are preferentially upregulated in rss3.Citation3 These include genes encoding biosynthetic enzymes for lignin, such as PAL, COMT, and CAD (). Lignin is a phenylpropanoid polymer derived from monolignols and a major component of the secondary cell wall,Citation9 where lignin is deposited between the space of cellulose microfibrils. Since RSS3 expresses and functions in the meristematic zone (MZ) and in the region that borders the MZ within the elongation zone (EZ) of the root tip,Citation3 we monitored lignin contents in the cell wall areas in the MZ, by measuring the UV absorbance (278 nm) of ultra-thin sections under a UV microscope.Citation10 As shown in , UV absorption in the cell wall areas was detected only at low levels in the wild type (WT), but at considerable levels in rss3. This indicates that lignin deposition into the cell walls in the MZ is restricted in WT, whereas it is ectopically induced in rss3. Because lignin confers rigidity to cell walls, lignin deposition may be a cause of the reduced cell elongation in rss3 under conditions without salinity. However, this would not account for the impaired cell elongation in the rss3 root under saline conditions, where the UV absorbance remained unchanged between WT and rss3 (). Lignin deposition appeared to be perturbed by yet unknown cellular responses caused by salt stress in rss3, in spite of activation of lignin synthetic gene expression. Expression of nearly half of salt stress-responsive genes is modified in rss3.Citation3 The largely altered salt stress responses are rather likely to be responsible for drastically impaired cell elongation in rss3 under the salinity conditions.
Figure 1. Altered expression of genes encoding lignin biosynthetic enzymes (A) and NAC family transcription factors (B) in the rss3 root tip grown in the absence and presence of 100 mM NaCl.Citation3 Genes encoding lignin biosynthetic enzymes or NAC transcription factors that were upregulated in rss3 were selected, based on the microarray data (GEO repository: GSE41442),Citation3 and levels of gene expression were visualized with TIGR Multi-experiment Viewer (MeV; http://www.tm4.org/mev/).
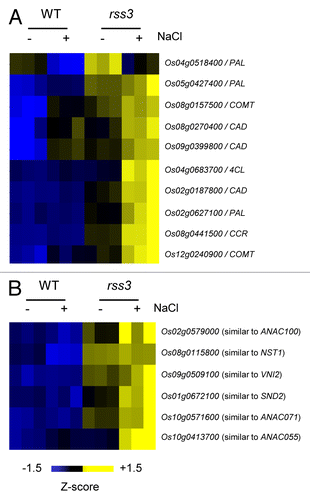
Figure 2. UV microspectrophotometry of cell walls in the root meristematic zone. (A) (first 4) Phenylpropanoid-derived compounds deposited in cell walls were observed in transverse sections (3 μm thickness) prepared from the meristematic zone of root tips (300 µm distal from the root cap tip) of 3-d-old wild-type and rss3 grown in the absence (−) and presence (+) of 100 mM NaCl. (A) (second 4) Schematic drawing of the position of cell walls in the sections shown in the first 4 panels. (B) Quantification of UV absorbance in the cell wall observed in (A). Root samples were fixed in 3% glutaraldehyde, treated with 50% hypochlorous acid for deproteinization, and stained using 1% osmium tetroxide. After dehydration in a graded ethanol series, the root tips were embedded in epoxy resin and sliced with a diamond knife. Sections were mounted on quartz microscope slides, covered with a quartz coverslip, and observed at a wavelength of 278 nm under a microspectrophotometer (Zeiss MPM800), with a bandwidth of 20 nm and objective lens magnification of × 40. The levels of UV absorption in the cell wall and cytosol (for the background) were quantified using ImageJ software (http://rsbweb.nih.gov/ij/). Measurements were taken from at least 4 different positions for each sample and averaged for calculation. Data of at least 3 biological replicates were used for each condition (Mean ± SD n ≥ 3). Asterisk indicates P value < 0.05 (the Student t-test).
It is known that lignin and cellulose biosynthesis is coordinately controlled. For example, chemical treatment or mutations that impair cellulose biosynthesis cause increased lignin deposition.Citation11-Citation15 Oppositely, downregulation of lignin biosynthetic pathways results in increased cellulose synthesis.Citation16 Therefore, it is plausible that the increased lignin content in rss3 may reflect the overall alteration of cell wall properties. To verify this possibility, we examined the effect of isoxaben, a cellulose synthase inhibitor, on root growth of WT and rss3. As shown in , root growth is impaired by isoxaben in WT and rss3 in dose-dependent manners. Noticeably, a low concentration of isoxaben (0.01 μM) inhibited root growth of rss3 but not that of WT (), indicating that rss3 is hypersensitive to the inhibitor. The inhibition by 0.01 μM isoxaben in rss3 was accompanied by reduced sizes in both MZ and EZ (). The size reduction was concomitant with a significant decrease in cell number of the MZ but not of the EZ (). This suggests that cell division is affected in the mutant by inhibiting cellulose synthase. In contrast, the size reduction in the EZ probably reflects that inhibition of cellulose synthase leads to a decrease in cell elongation in the rss3 root tip. The hypersensitivity of rss3 to the cellulose synthase inhibitor implies that drastic changes in characteristics of cell walls are induced when cellulose synthesis is inhibited in rss3, where some unknown changes in cell walls have already occurred.
Figure 3. Sensitivity of the roots of wild-type and rss3 to isoxaben. (A) Dose-dependent effects of isoxaben on root growth. Mean ± SD, n > 4. (B) Effects of isoxaben on the size and number of cells in the meristematic zone (MZ) and elongation zone (EZ) of the root tip. Mean ± SD, n = 3. Asterisks indicate the Student t-test P value < 0.05. Surface-sterilized seeds were germinated on an agar-based medium [agar 0.8%, 1 mM KH2PO4, 0.05% MES-KOH (pH 5.8)] containing 0.01 μM, 0.05 μM, and 0.1 μM isoxaben or mock solution (1 × 10−4% DMSO in final) in a rectangular petri dish to treat rice seedlings with isoxaben (Wako, 092–05961). Seedlings were grown on the medium in plates tilted at a 75° angle.
![Figure 3. Sensitivity of the roots of wild-type and rss3 to isoxaben. (A) Dose-dependent effects of isoxaben on root growth. Mean ± SD, n > 4. (B) Effects of isoxaben on the size and number of cells in the meristematic zone (MZ) and elongation zone (EZ) of the root tip. Mean ± SD, n = 3. Asterisks indicate the Student t-test P value < 0.05. Surface-sterilized seeds were germinated on an agar-based medium [agar 0.8%, 1 mM KH2PO4, 0.05% MES-KOH (pH 5.8)] containing 0.01 μM, 0.05 μM, and 0.1 μM isoxaben or mock solution (1 × 10−4% DMSO in final) in a rectangular petri dish to treat rice seedlings with isoxaben (Wako, 092–05961). Seedlings were grown on the medium in plates tilted at a 75° angle.](/cms/asset/1bc4dab1-50f9-4184-9846-f826a4cb3cb1/kpsb_a_10926256_f0003.gif)
There have been cumulating lines of evidence that interference with cell wall integrity causes increased jasmonate production and ectopic lignin deposition that is mediated by jasmonate-dependent and -independent mechanisms.Citation11-Citation13 Therefore, the hypersensitivity to isoxaben may also be explained by that jasmonate signaling provoked by inhibition of cellulose synthase is enhanced in rss3. This seems consistent with our observation that jasmonate-responsive genes are excessively induced in rss3,Citation3 and the report that accumulation of monolignol is induced by jasmonate.Citation17 To the contrary, however, it was reported that cell wall damage-induced lignin deposition was increased by mutations in the jasmonate signaling pathway at least in the EZ of the root tip in Arabidopsis.Citation14,Citation15 Future research is needed to determine which components other than lignin are affected in the cell walls in rss3, and how overall properties of cell walls are controlled in the MZ.
The regulation of genes encoding enzymes for cell wall metabolism by RSS3 and its binding proteins may be mediated by secondary wall master regulators. In Arabidopsis, MYB-type (MYB46, MYB58, and MYB63) and NAC-type (SND1, SND2, VND6, VND7, NST1, and NST2) transcription factors function as positive regulators of secondary cell wall synthesis.Citation18-Citation24 Some rice NAC-type transcription factors have also been described to control secondary cell wall metabolism.Citation25 In rss3, several NAC genes that belong to the subfamily, in which Arabidopsis NST1 and SND2 are classified, were upregulated (). Thus, these factors may participate in the control of cell wall metabolisms in the MZ in the root apex. Interestingly, it was reported that lignin and cellulose synthetic genes are oppositely regulated in rice.Citation26 In contrast, however, we could not find preferential downregulation of cellulose synthetic genes in the rss3 root tip in our microarray analysis.Citation3
Mutants with altered cell wall composition exhibit defects in root elongation that are enhanced under stress conditions. For example, mutation of the chitinase-like protein AtCTL1 causes ectopic deposition of lignin, aberrant cell shape, and hyper-sensitivity to heat, salt, and dehydration stresses.Citation27-Citation29 Furthermore, cellulose-deficient mutants rsw1 and rsw2 are sensitive to heat and salt stress.Citation6,Citation30 Therefore, cell wall integrity is likely important for both root elongation and stress tolerance. Changes in the cell wall properties other than lignin deposition may also be a cause of hypersensitivity to salt stress in rss3.
In conclusion, we propose that RSS3, which binds to the JAZ and class-C bHLH factors, has a role to modulate cell wall properties. Together with the observation that the RSS3 complex functions to sustain root growth by preventing an excessive jasmonic acid response at the root apex,Citation3 the regulation mediated by RSS3 might be important for coordinated control between root elongation and cell wall plasticity under particular environmental stress conditions.
| Abbreviations: | ||
| RSS3 | = | RICE SALT SENSITIVE3 |
| JAZ | = | JASMONATE ZIM DOMAIN, bHLH, basic helix-loop-helix |
| PAL | = | PHENYLALANINE AMMONIA LYASE |
| COMT | = | caffeic acid-O-methyltransferase |
| CAD | = | cinnamyl alcohol dehydrogenase |
| CCR | = | Cinnamoyl-CoA reductase |
| MZ | = | meristematic zone |
| EZ | = | elongation zone |
Acknowledgments
We thank Taisuke Nishimura and Ken-ichi Kurotani for helpful discussion. This work was supported in part by JSPS KAKENHI (24580493).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Iraki NM, Singh N, Bressan RA, Carpita NC. Cell walls of tobacco cells and changes in composition associated with reduced growth upon adaptation to water and saline stress. Plant Physiol 1989; 91:48 - 53; http://dx.doi.org/10.1104/pp.91.1.48; PMID: 16667041
- Cosgrove DJ. Assembly and enlargement of the primary cell wall in plants. Annu Rev Cell Dev Biol 1997; 13:171 - 201; http://dx.doi.org/10.1146/annurev.cellbio.13.1.171; PMID: 9442872
- Toda Y, Tanaka M, Ogawa D, Kurata K, Kurotani K, Habu Y, Ando T, Sugimoto K, Mitsuda N, Katoh E, et al. RICE SALT SENSITIVE3 forms a ternary complex with JAZ and class-C bHLH factors and regulates jasmonate-induced gene expression and root cell elongation. Plant Cell 2013; 25:1709 - 25; http://dx.doi.org/10.1105/tpc.113.112052; PMID: 23715469
- Ogawa D, Abe K, Miyao A, Kojima M, Sakakibara H, Mizutani M, Morita H, Toda Y, Hobo T, Sato Y, et al. RSS1 regulates the cell cycle and maintains meristematic activity under stress conditions in rice. Nat Commun 2011; 2:278; http://dx.doi.org/10.1038/ncomms1279; PMID: 21505434
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R, et al. Molecular analysis of cellulose biosynthesis in Arabidopsis.. Science 1998; 279:717 - 20; http://dx.doi.org/10.1126/science.279.5351.717; PMID: 9445479
- Lane DR, Wiedemeier A, Peng L, Höfte H, Vernhettes S, Desprez T, Hocart CH, Birch RJ, Baskin TI, Burn JE, et al. Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-beta-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol 2001; 126:278 - 88; http://dx.doi.org/10.1104/pp.126.1.278; PMID: 11351091
- Roudier F, Fernandez AG, Fujita M, Himmelspach R, Borner GH, Schindelman G, Song S, Baskin TI, Dupree P, Wasteneys GO, et al. COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 2005; 17:1749 - 63; http://dx.doi.org/10.1105/tpc.105.031732; PMID: 15849274
- Tsang DL, Edmond C, Harrington JL, Nühse TS. Cell wall integrity controls root elongation via a general 1-aminocyclopropane-1-carboxylic acid-dependent, ethylene-independent pathway. Plant Physiol 2011; 156:596 - 604; http://dx.doi.org/10.1104/pp.111.175372; PMID: 21508182
- Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu Rev Plant Biol 2003; 54:519 - 46; http://dx.doi.org/10.1146/annurev.arplant.54.031902.134938; PMID: 14503002
- Yoshida M, Ohta H, Yamamoto H, Okuyama T. Tensile growth stress and lignin distribution in the cell walls of yellow poplar, Liriodendron tulipifera Linn. Trees (Berl) 2002; 16:457 - 64; http://dx.doi.org/10.1007/s00468-002-0186-2
- Ellis C, Turner JG. The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 2001; 13:1025 - 33; http://dx.doi.org/10.1105/tpc.13.5.1025; PMID: 11340179
- Ellis C, Karafyllidis I, Wasternack C, Turner JG. The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 2002; 14:1557 - 66; http://dx.doi.org/10.1105/tpc.002022; PMID: 12119374
- Caño-Delgado A, Penfield S, Smith C, Catley M, Bevan M. Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana.. Plant J 2003; 34:351 - 62; http://dx.doi.org/10.1046/j.1365-313X.2003.01729.x; PMID: 12713541
- Hamann T, Bennett M, Mansfield J, Somerville C. Identification of cell-wall stress as a hexose-dependent and osmosensitive regulator of plant responses. Plant J 2009; 57:1015 - 26; http://dx.doi.org/10.1111/j.1365-313X.2008.03744.x; PMID: 19036034
- Denness L, McKenna JF, Segonzac C, Wormit A, Madhou P, Bennett M, Mansfield J, Zipfel C, Hamann T. Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in Arabidopsis.. Plant Physiol 2011; 156:1364 - 74; http://dx.doi.org/10.1104/pp.111.175737; PMID: 21546454
- Li L, Zhou Y, Cheng X, Sun J, Marita JM, Ralph J, Chiang VL. Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc Natl Acad Sci U S A 2003; 100:4939 - 44; http://dx.doi.org/10.1073/pnas.0831166100; PMID: 12668766
- Pauwels L, Morreel K, De Witte E, Lammertyn F, Van Montagu M, Boerjan W, Inzé D, Goossens A. Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc Natl Acad Sci U S A 2008; 105:1380 - 5; http://dx.doi.org/10.1073/pnas.0711203105; PMID: 18216250
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 2005; 19:1855 - 60; http://dx.doi.org/10.1101/gad.1331305; PMID: 16103214
- Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 2005; 17:2993 - 3006; http://dx.doi.org/10.1105/tpc.105.036004; PMID: 16214898
- Zhong R, Demura T, Ye ZH. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis.. Plant Cell 2006; 18:3158 - 70; http://dx.doi.org/10.1105/tpc.106.047399; PMID: 17114348
- Zhong R, Richardson EA, Ye ZH. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis.. Plant Cell 2007; 19:2776 - 92; http://dx.doi.org/10.1105/tpc.107.053678; PMID: 17890373
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis.. Plant Cell 2008; 20:2763 - 82; http://dx.doi.org/10.1105/tpc.108.061325; PMID: 18952777
- Zhou J, Lee C, Zhong R, Ye ZH. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis.. Plant Cell 2009; 21:248 - 66; http://dx.doi.org/10.1105/tpc.108.063321; PMID: 19122102
- Ohashi-Ito K, Oda Y, Fukuda H. Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 2010; 22:3461 - 73; http://dx.doi.org/10.1105/tpc.110.075036; PMID: 20952636
- Zhong R, Lee C, McCarthy RL, Reeves CK, Jones EG, Ye ZH. Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol 2011; 52:1856 - 71; http://dx.doi.org/10.1093/pcp/pcr123; PMID: 21908441
- Ambavaram MM, Krishnan A, Trijatmiko KR, Pereira A. Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiol 2011; 155:916 - 31; http://dx.doi.org/10.1104/pp.110.168641; PMID: 21205614
- Zhong R, Kays SJ, Schroeder BP, Ye ZH. Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell 2002; 14:165 - 79; http://dx.doi.org/10.1105/tpc.010278; PMID: 11826306
- Mouille G, Robin S, Lecomte M, Pagant S, Höfte H. Classification and identification of Arabidopsis cell wall mutants using Fourier-Transform InfraRed (FT-IR) microspectroscopy. Plant J 2003; 35:393 - 404; http://dx.doi.org/10.1046/j.1365-313X.2003.01807.x; PMID: 12887590
- Kwon Y, Kim SH, Jung MS, Kim MS, Oh JE, Ju HW, Kim KI, Vierling E, Lee H, Hong SW. Arabidopsis hot2 encodes an endochitinase-like protein that is essential for tolerance to heat, salt and drought stresses. Plant J 2007; 49:184 - 93; http://dx.doi.org/10.1111/j.1365-313X.2006.02950.x; PMID: 17156413
- Kang JS, Frank J, Kang CH, Kajiura H, Vikram M, Ueda A, Kim S, Bahk JD, Triplett B, Fujiyama K, et al. Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc Natl Acad Sci U S A 2008; 105:5933 - 8; http://dx.doi.org/10.1073/pnas.0800237105; PMID: 18408158