Abstract
Asian soybean rust (ASR) caused by the fungus Phakopsora pachyrhizi is one of the most devastating foliar diseases affecting soybean production worldwide. Even though several resistance sources have been identified in soybean, they do not show resistance to all races of P. pachyrhizi. Identification of genes that confer nonhost resistance (NHR) against P. pachyrhizi in another legume species will provide an avenue to engineer soybean to have durable and broad spectrum resistance against P. pachyrhizi strains. Recently, we identified a Medicago truncatula gene, IRG1 (INHIBITOR OF RUST GERM-TUBE DIFFERENTIATION1), that when mutated inhibits the growth of P. pachyrhizi. IRG1 encodes a Cys(2)His(2) zinc finger transcription factor that controls wax-biosynthesis-related genes. The irg1 mutant shows a complete loss of abaxial epicuticular wax crystals and surface hydrophobicity, resulting in the inhibition of pre-penetration structure formation. In order to confirm the role of surface hydrophobicity in the formation of pre-penetration structures, we examined the expression profiles of P. pachyrhizi putative pre-penetration structure-development-related genes on a solid surface or a M. truncatula abaxial leaf surface. Interestingly, the expression of kinase family genes was upregulated on the hydrophobic surface and M. truncatula wild-type leaf surface, but not on the M. truncatula irg1 mutant leaf surface, suggesting that these genes play a role in P. pachyrhizi pre-penetration structure development. In addition, our results suggest that hydrophobicity on the M. truncatula leaf surface may function as a key signal to induce the P. pachyrhizi genes involved in pre-penetration structure development.
Asian soybean rust (ASR) caused by biotrophic plant pathogenic fungus, Phakopsora pachyrhizi is one of the devastating diseases of soybean. The disease cycle of P. pachyrhizi begins with urediniospores, which have an important role in the disease cycle. The urediniospores attach to the surface of host leaves and produce pre-penetration structures, including germ tubes and appressoria. Unlike other rust pathogens, P. pachyrhizi is a unique, directly penetrating rust fungus. After penetration, P. pachyrhizi develops infection hyphae, colonizes host cells, and forms a specialized feeding structure called haustorium. P. pachyrhizi develops tan lesions on the leaf surface of a susceptible soybean plant one week after infection and then makes uredinia, which are structures that produce urediniospores on the abaxial leaf surface.Citation1,Citation2 Five soybean resistance genes, Rpp1–5, confer immunity or resistance to P. pachyrhizi.Citation3-Citation6 However, there is no soybean line that has broad-spectrum disease resistance to all races of P. pachyrhizi.Citation7 Therefore, the demand for development of durable resistance to P. pachyrhizi is high. Understanding the mechanism of plant immunity against P. pachyrhizi would benefit the development of durable resistant plants. The nonhost resistance (NHR) is the most common and durable form of resistance against potential pathogens in nature. NHR mechanisms can be utilized for improving resistance to pathogen infection in crop plants.Citation8-Citation10
Medicago truncatula, a model plant species for legumes, shows NHR response to P. pachyrhizi. It has been demonstrated that P. pachyrhizi forms germ tubes with appressoria and penetrates into epidermal cells, resulting in necrotic symptoms without sporulation on M. truncatula.Citation11 To identify mutants that show an altered phenotype to P. pachyrhizi infection, a forward genetics screen using Tnt1 insertion mutant lines of M. truncatulaCitation12 has been developed and an inhibitor of rust germ-tube differentiation1 (irg1) mutant that inhibited fungal pre-penetration structure differentiation was identified.Citation11 Interestingly, irg1 mutants showed pre-penetration resistance against rust pathogens, including P. pachyrhizi and Puccinia emaculata (switchgrass pathogen), and the hemibiotrophic anthracnose fungus Colletotrichum trifolii, but not to necrotrophic fungal pathogens Phoma medicagenis and Sclerotinia sclerotiorum.Citation11 Leaves of irg1 mutant lack abaxial epicuticular wax crystals, indicating that inhibition of rust pre-infection structures in irg1 mutant is connected with the loss of surface hydrophobicity. Furthermore, we demonstrated that IRG1 encodes the Cys(2)His(2) zinc finger type transcription factor that regulates wax biosynthetic pathways in M. truncatula.Citation11 To investigate the function of epicuticular waxes in stimulating the differentiation of fungal pre-penetration structures such as germ tubes and appressoria, we performed quantitative analyses of fungal development during in vitro germination assay. First, epicuticular waxes were extracted with hexane. Then, urediniospores of P. pachyrhizi were put on hydrophilic glass surfaces coated with or without epicuticular waxes isolated from both surfaces of M. truncatula wild type and irg1 mutants, and kept in a high humidity chamber. Although the waxes isolated from the adaxial leaf surface of both wild-type and irg1 mutant induced the formation of pre-penetration structures compared with the mock (hexane-coated slide glass), there was no significant difference in their ability to induce the pre-penetration structures between wild-type and irg1 mutants. However, we found a significant reduction in the formation of pre-penetration structures on the glass slides coated with waxes isolated from the abaxial leaf surface of irg1 mutants compared with wild-type, suggesting that epicuticular waxes or hydrophobicity promote the formation of pre-penetration structures such as germ tubes and appressoria.Citation11
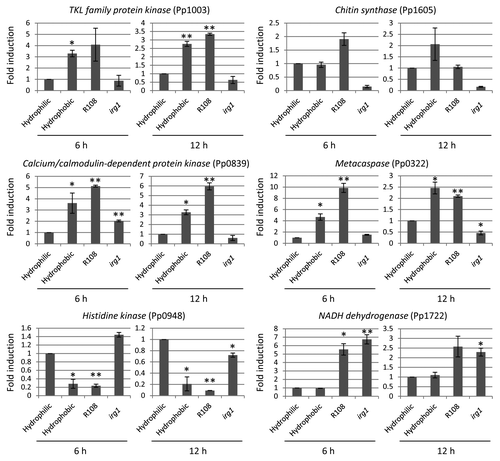
The formation of pre-penetration structures is a crucial step for the pathogenicity of rust pathogens including P. pachyrhizi.Citation1 Therefore, understanding the molecular basis of mechanisms related to the formation of pre-penetration structures is essential for providing novel strategies for disease management. In other economically important fungal pathogens such as rice blast, several signaling pathways including mitogen-activated protein kinase (MAPK) signaling pathways, G-protein-mediated signaling pathways, calcium/calmodulin-mediated signaling pathways, and cAMP-mediated signaling pathways have been identified. Mutant analyses of genes involved in these signaling pathways have revealed that these pathways are required not only for the formation of pre-penetration structures, but also for the pathogenicity of rice blast.Citation13 Unlike rice blast, limited information is available for the regulators of pre-penetration structure development on rust pathogens. In order to investigate the regulation of pre-penetration structure development in P. pachyrhizi, we examined the expression profiles of P. pachyrhizi pre-penetration structure-development-related genes on a solid surface or M. truncatula abaxial leaf surface. We selected putative pre-penetration structure-development-related genes, including chitin synthase, kinase family gene, and metacaspase, and housekeeping gene, NADH dehydrogenase (Pp1722) from P. pachyrhizi expressed sequence tag,Citation7 and performed RT-qPCR analysis using gene-specific primer sets (). It is interesting that the abaxial leaf surface of the irg1 mutant showed reduced pre-penetration structure development of P. pachyrhizi.Citation11 Therefore, one could argue that the regulation of pre-penetration structure-development-related genes on the irg1 mutant may result from reduced viability of urediniospores rather than from the direct effects of host signals such as epicuticular waxes or hydrophobicity. To rule out this possibility, we investigated the expression of NADH dehydrogenase (Pp1722), and found no significant difference on the gene expression between M. truncatula wild-type and the irg1 mutant. The expression of kinase family genes, including TKL family protein kinase (Pp1003) and calcium/calmodulin-dependent protein kinase (Pp0839), chitin synthase (Pp1605), and metacaspase (Pp0322) was upregulated on the hydrophobic surface and M. truncatula wild-type leaf surface, but not on the M. truncatula irg1 mutant leaf surface, suggesting that these genes may have a role in pre-penetration structure development in response to epicuticular waxes or hydrophobicity (). It has been demonstrated that the calcium/calmodulin-mediated signaling pathway is involved not only in fundamental physiological processes, but also in the pathogenicity of filamentous fungi.Citation14 It was reported that the expression of calcium/calmodulin-dependent protein kinase was upregulated in response to the hard surface contact in rice blast and anthracnose pathogens.Citation15,Citation16 These results suggest that P. pachyrhizi also preserve the calcium/calmodulin-mediated signaling pathway to regulate pre-penetration structure development. The expression of histidine kinase (Pp0948) was downregulated on the hydrophobic surface and M. truncatula wild-type leaf surface compared with the M. truncatula irg1 mutant leaf surface, suggesting that histidine kinase may have negative functions on pre-penetration structure development. Histidine kinases are known as important mediators for adaptation to stresses from prokaryotes to eukaryotes.Citation17,Citation18 In addition, histidine kinases have been demonstrated to have an impact on the pathogenicity of fungal pathogens such as Botrytis cinerea, Fusarium oxysporum, Claviceps purpurea, and rice blast.Citation19-Citation22 Interestingly, histidine kinase was reported to impact the pathogenicity of Alternaria longipes. It has been demonstrated that the A. longipes histidine kinase (AlHK1)-null mutant produced larger lesions compared with the wild-type strain, suggesting that histidine kinase functions as a negative regulator for the pathogenicity of A. longipes.Citation23 Taken together, histidine kinase (Pp0948) may be involved in the pathogenicity of P. pachyrhizi in a negative manner. These results indicate that epicuticular waxes or hydrophobicity may function as key signals for the pre-penetration structure development of P. pachyrhizi.
Table 1. List of gene-specific primer sets for RT-qPCR of P. pachyrhizi
Figure 1. Expression profiles of TKL family protein kinase (Pp1003), chitin synthase (Pp1605), calcium/calmodulin-dependent protein kinase (Pp0839), metacaspase (Pp0322), histidine kinase (Pp0948), and NADH dehydrogenase (Pp1722) during pre-penetration structure development of P. pachyrhizi. Fresh urediniospores (1 × 105 spores/ml) were incubated on hydrophilic glass plates and hydrophobic petri plates, or spot-inoculated on M. truncatula wild-type (R108) and irg1 mutants. The inoculated plates and leaves were incubated in the dark, and then total RNA was isolated using TRIzol® reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Five μg of DNase-treated RNA was reverse transcribed, and the cDNA (1:10) was then used for RT-qPCR. The average threshold cycle (CT) values calculated from triplicate biological samples were used to determine the fold expression relative to the controls. Primers specific for elongation factor 1α (Pp1107) and ubiquitin 5 (Pp1724) were used to normalize differences in template amounts. Vertical bars indicate the standard errors for 3 independent experiments. Asterisks indicate a significant difference from hydrophilic glass plate using a t-test (* = p < 0.05, ** = p < 0.01).
In conclusion, we demonstrated the importance of host signals such as epicuticular waxes and/or hydrophobicity in the pre-penetration structure development of P. pachyrhizi. Although the precise role of IRG1 in wax biosynthesis and the mechanism by which hydrophobicity induces fungal genes involved in pathogenicity need further investigation, our results present the possibility of a novel strategy for disease management by manipulating plant surface hydrophobicity.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by a grant from BASF Plant Sciences, Germany, and The Samuel Roberts Noble Foundation.
References
- Goellner K, Loehrer M, Langenbach C, Conrath U, Koch E, Schaffrath U. Phakopsora pachyrhizi, the causal agent of Asian soybean rust. Mol Plant Pathol 2010; 11:169 - 77; http://dx.doi.org/10.1111/j.1364-3703.2009.00589.x; PMID: 20447267
- Loehrer M, Langenbach C, Goellner K, Conrath U, Schaffrath U. Characterization of nonhost resistance of Arabidopsis to the Asian soybean rust. Mol Plant Microbe Interact 2008; 21:1421 - 30; http://dx.doi.org/10.1094/MPMI-21-11-1421; PMID: 18842092
- Hyten DL, et al. Map Location of the Locus That Confers Resistance to Soybean Rust in Soybean. Crop Sci 2007; 47:837 - 8; http://dx.doi.org/10.2135/cropsci2006.07.0484
- Monteros M, Ha B-K, Phillips D, Boerma H. SNP assay to detect the ‘Hyuuga’ red-brown lesion resistance gene for Asian soybean rust. TAG Theoretical and Applied Genetics 2010; 121:1023 - 32; http://dx.doi.org/10.1007/s00122-010-1368-8
- Garcia A, Calvo ES, de Souza Kiihl RA, Harada A, Hiromoto DM, Vieira LG. Molecular mapping of soybean rust (Phakopsora pachyrhizi) resistance genes: discovery of a novel locus and alleles. Theor Appl Genet 2008; 117:545 - 53; http://dx.doi.org/10.1007/s00122-008-0798-z; PMID: 18506417
- Silva DC, Yamanaka N, Brogin RL, Arias CA, Nepomuceno AL, Di Mauro AO, Pereira SS, Nogueira LM, Passianotto AL, Abdelnoor RV. Molecular mapping of two loci that confer resistance to Asian rust in soybean. Theor Appl Genet 2008; 117:57 - 63; http://dx.doi.org/10.1007/s00122-008-0752-0; PMID: 18392802
- Posada-Buitrago ML, Frederick RD. Expressed sequence tag analysis of the soybean rust pathogen Phakopsora pachyrhizi. Fungal Genet Biol 2005; 42:949 - 62; http://dx.doi.org/10.1016/j.fgb.2005.06.004; PMID: 16291502
- Mysore KS, Ryu CM. Nonhost resistance: how much do we know?. Trends Plant Sci 2004; 9:97 - 104; http://dx.doi.org/10.1016/j.tplants.2003.12.005; PMID: 15102376
- Fan J, Doerner P. Genetic and molecular basis of nonhost disease resistance: complex, yes; silver bullet, no. Curr Opin Plant Biol 2012; 15:400 - 6; http://dx.doi.org/10.1016/j.pbi.2012.03.001; PMID: 22445191
- Heath MC. Nonhost resistance and nonspecific plant defenses. Curr Opin Plant Biol 2000; 3:315 - 9; http://dx.doi.org/10.1016/S1369-5266(00)00087-X; PMID: 10873843
- Uppalapati SR, Ishiga Y, Doraiswamy V, Bedair M, Mittal S, Chen J, Nakashima J, Tang Y, Tadege M, Ratet P, et al. Loss of abaxial leaf epicuticular wax in Medicago truncatula irg1/palm1 mutants results in reduced spore differentiation of anthracnose and nonhost rust pathogens. Plant Cell 2012; 24:353 - 70; http://dx.doi.org/10.1105/tpc.111.093104; PMID: 22294617
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, et al. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J 2008; 54:335 - 47; http://dx.doi.org/10.1111/j.1365-313X.2008.03418.x; PMID: 18208518
- Ebbole DJ. Magnaporthe as a model for understanding host-pathogen interactions. Annu Rev Phytopathol 2007; 45:437 - 56; http://dx.doi.org/10.1146/annurev.phyto.45.062806.094346; PMID: 17489691
- Lengeler KB, Davidson RC, D’souza C, Harashima T, Shen WC, Wang P, Pan X, Waugh M, Heitman J. Signal transduction cascades regulating fungal development and virulence. Microbiol Mol Biol Rev 2000; 64:746 - 85; http://dx.doi.org/10.1128/MMBR.64.4.746-785.2000; PMID: 11104818
- Kim YK, Li D, Kolattukudy PE. Induction of Ca2+-calmodulin signaling by hard-surface contact primes Colletotrichum gloeosporioides conidia to germinate and form appressoria. J Bacteriol 1998; 180:5144 - 50; PMID: 9748448
- Liu ZM, Kolattukudy PE. Early expression of the calmodulin gene, which precedes appressorium formation in Magnaporthe grisea, is inhibited by self-inhibitors and requires surface attachment. J Bacteriol 1999; 181:3571 - 7; PMID: 10348871
- Schaller GE, Shiu SH, Armitage JP. Two-component systems and their co-option for eukaryotic signal transduction. Curr Biol 2011; 21:R320 - 30; http://dx.doi.org/10.1016/j.cub.2011.02.045; PMID: 21549954
- Jung K, Fried L, Behr S, Heermann R. Histidine kinases and response regulators in networks. Curr Opin Microbiol 2012; 15:118 - 24; http://dx.doi.org/10.1016/j.mib.2011.11.009; PMID: 22172627
- Viaud M, Fillinger S, Liu W, Polepalli JS, Le Pêcheur P, Kunduru AR, Leroux P, Legendre L. A class III histidine kinase acts as a novel virulence factor in Botrytis cinerea. Mol Plant Microbe Interact 2006; 19:1042 - 50; http://dx.doi.org/10.1094/MPMI-19-1042; PMID: 16941908
- Rispail N, Di Pietro A. The two-component histidine kinase Fhk1 controls stress adaptation and virulence of Fusarium oxysporum. Mol Plant Pathol 2010; 11:395 - 407; http://dx.doi.org/10.1111/j.1364-3703.2010.00612.x; PMID: 20447287
- Nathues E, Jörgens C, Lorenz N, Tudzynski P. The histidine kinase CpHK2 has impact on spore germination, oxidative stress and fungicide resistance, and virulence of the ergot fungus Claviceps purpurea. Mol Plant Pathol 2007; 8:653 - 65; http://dx.doi.org/10.1111/j.1364-3703.2007.00421.x; PMID: 20507528
- Zhang HF, Liu K, Zhang X, Song W, Zhao Q, Dong Y, Guo M, Zheng X, Zhang Z. A two-component histidine kinase, MoSLN1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Curr Genet 2010; 56:517 - 28; http://dx.doi.org/10.1007/s00294-010-0319-x; PMID: 20848286
- Luo YY, Yang JK, Zhu ML, Liu CJ, Li HY, Lu ZB, Pan WZ, Zhang ZH, Bi W, Zhang KQ. The group III two-component histidine kinase AlHK1 is involved in fungicides resistance, osmosensitivity, spore production and impacts negatively pathogenicity in Alternaria longipes. Curr Microbiol 2012; 64:449 - 56; http://dx.doi.org/10.1007/s00284-012-0093-8; PMID: 22349956