Abstract
Verticillium longisporum is a soil-borne vascular pathogen found primarily on oilseed rape in Northern Europe. Infection of the model plant Arabidopsis thaliana can be achieved under laboratory conditions. In the article related to this addendum, we have shown that Arabidopsis dde2–2 mutants that are compromised in their ability to synthesize the defense hormone jasmonoyl-isoleucine (JA-Ile) are slightly more susceptible than wild-type. Contrary to the expectation that hormone biosynthesis mutants and their respective receptor mutants should have the same phenotype, we found that plants that lack the JA-Ile receptor CORONATINE INSENSITIVE1 (COI1) are more tolerant to the disease. This addendum addressed the question whether the increased JA-Ile levels found in coi1 are responsible for its tolerance phenotype. Based on the evidence that the JA-Ile-deficient dde2–2 coi1-t double mutant is as tolerant as coi1-t, we conclude that increased JA-Ile levels do not protect Arabidopsis against the fungus in the absence of COI1.
Vascular pathogens like Fusarium oxysporum and Verticillium spp. enter their hosts through the roots and colonize the xylem vessels throughout the plant. Plant defense responses that are elicited through the main defense hormones jasmonoyl-isoleucine (JA-Ile) or salicylic acid (SA) do not have a major impact on the susceptibility of Arabidopsis against F. oxysporum and V. longisporum.Citation1,Citation2 In contrast, the coi1 mutant, which is compromised in JA-Ile signaling,Citation3,Citation4 is more tolerant against both fungi. Although initial colonization of the roots by either F. oxysporum or V. longisporum is not compromised in coi1, symptom development and fungal proliferation in the aerial part are less pronounced. This tolerance is not due to the hyper-activation of the SA pathway in both pathosystems. Moreover, grafting studies had revealed that the susceptibility-enhancing COI1 function acts in the roots. In summary, these results have unraveled a novel COI1 function in the roots, which is important for a yet unknown root-to-shoot signaling process that enables F. oxysporum and V. longisporum to elicit disease symptoms in shoots. Here, we addressed the importance of JA-Ile for the coi1 phenotype.
In our previous study, we have analyzed plant defense hormone levels in petioles of Arabidopsis plants, where the fungus predominantly completes its life cycle.Citation1 We have shown that petioles of the coi1-t mutant contain 6-fold increased basal levels of JA-Ile.Citation1 After 15 d post inoculation of up-rooted plants, JA-Ile levels increased in wild-type by a factor of 6 and in coi1-t by a factor of 2.5. Thus, 2.5-fold higher JA-Ile levels are present in infected coi1 plants as compared with infected wild-type. This raised the question whether JA-Ile might be responsible for the tolerance of coi1 against V. longisporum. Indeed, genes involved in the detoxification response like glutathione S-transferases GSTU1 and GSTU7 and transcription factor NAC032 are induced to higher levels by JA in coi1 than in wild-type,Citation5 suggesting that JA and/or its derivatives might act as xenobiotics if they are not properly perceived by a receptor.
JA-Ile is not responsible for the tolerance of the Arabidopsis coi1 mutant against Verticillium longisporum
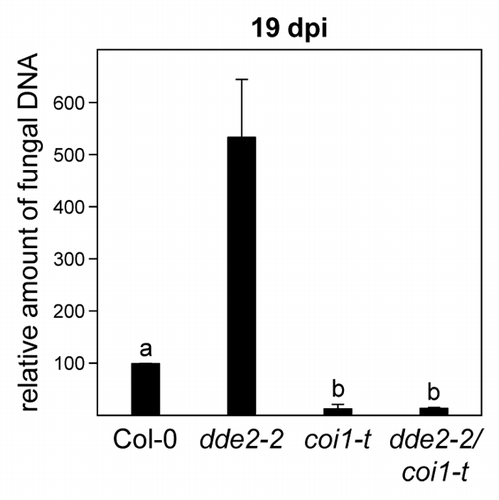
In order to address the question of whether increased JA-Ile levels are responsible for the observed coi1-mediated tolerance against V. longisporum, we infected the previously described dde2–2 coi1-t double mutantCitation5 side by side with wild-type plants and the corresponding single mutants and quantified disease symptoms by measuring the projected leaf area (). As described before,Citation1 plant growth was reduced to a similar extent in infected wild-type and dde2–2 plants, resulting in a decrease of projected leaf area by 50%. This effect was less pronounced in coi1-t plants. Importantly, the coi1-mediated tolerance was not altered by the dde2–2 allele. These data are supported by measurements of fungal DNA (). The fungus proliferated more vigorously in the dde2–2 mutant, indicating that JA-Ile-induced defense responses limit fungal growth to a certain degree in the presence of a functional COI1 allele. In contrast, fungal DNA levels were reduced in the coi1-t single and the dde2–2 coi1-t double mutant. It is concluded that jasmonate-induced processes are not the reason for the observed tolerance of coi1 against V. longisporum.
Figure 1. Disease phenotype of Verticillium longisporum-infected wild-type, dde2–2, coi1-t, and dde2–2 coi1-t plants at 19 d post inoculation. Relative leaf area of mock-infected and V. longisporum (V.l.)-infected plants of the indicated genotypes. Data are means (+/− SEM) of 68–70 replicates from 4 independent experiments. For each experiment and genotype, the mean leaf area of mock-infected plants was set to 100%, respectively. Different letters indicate significant differences at P < 0.001 (one-way ANOVA followed by Tukey-Kramer multiple comparison test) between V. longisporum-infected plants. Inoculation procedures and genotypes are described in Ralhan et al. 2012Citation1 and Köster et al. 2012.Citation5
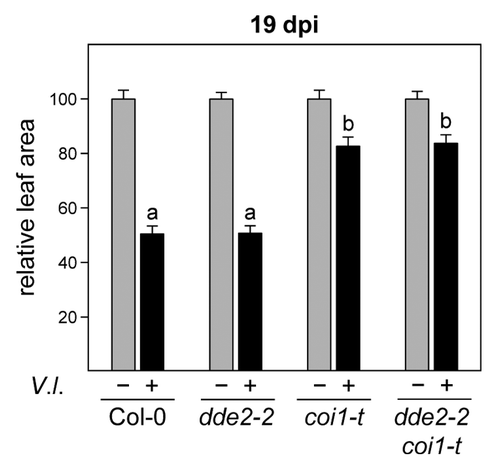
Figure 2. Fungal biomass of Verticillium longisporum-infected wild-type, dde2–2, coi1-t, and dde2–2 coi1-t plants. Relative quantification of fungal biomass by real-time PCR on DNA extracted from petioles of V. longisporum-infected wild-type, dde2–2, coi1-t, and dde2–2 coi1-t plants at 19 d post inoculation (dpi). Amplification values for fungal internal ribosomal spacer regions were normalized to the abundance of Arabidopsis Actin8 sequences. Relative amounts of fungal DNA were set to 100% for the wild-type. Bars indicate means (+/− SEM) of 3 to 4 biological replicates. Each replicate represents a pool of 4 plants. Different letters indicate significant differences at P < 0.001 (one-way ANOVA followed by Tukey-Kramer multiple comparison test) between V. longisporum-infected Col-0, coi1-t, and dde2–2 coi1-t plants. Inoculation procedures, determination of fungal DNA, and genotypes are described in Ralhan et al. 2012Citation1 and Köster et al. 2012.Citation5
| Abbreviations: | ||
| COI1 | = | CORONATINE INSENSITIVE1 |
| JA-Ile | = | jasmonoyl-isoleucine |
| SA | = | salicylic acid |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Ronald Scholz (University Göttingen) for excellent technical assistance.
References
- Ralhan A, Schöttle S, Thurow C, Iven T, Feussner I, Polle A, Gatz C. The vascular pathogen Verticillium longisporum requires a jasmonic acid-independent COI1 function in roots to elicit disease symptoms in Arabidopsis shoots. Plant Physiol 2012; 159:1192 - 203; http://dx.doi.org/10.1104/pp.112.198598; PMID: 22635114
- Thatcher LF, Manners JM, Kazan K. Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis.. Plant J 2009; 58:927 - 39; http://dx.doi.org/10.1111/j.1365-313X.2009.03831.x; PMID: 19220788
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007; 448:661 - 5; http://dx.doi.org/10.1038/nature05960; PMID: 17637677
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007; 448:666 - 71; http://dx.doi.org/10.1038/nature06006; PMID: 17637675
- Köster J, Thurow C, Kruse K, Meier A, Iven T, Feussner I, Gatz C. Xenobiotic- and jasmonic acid-inducible signal transduction pathways have become interdependent at the Arabidopsis CYP81D11 promoter. Plant Physiol 2012; 159:391 - 402; http://dx.doi.org/10.1104/pp.112.194274; PMID: 22452854