Abstract
In a recent addendum, Oren Tzfadia and Gad Galili (PSB 2014; 9:e26732) showed that several Arabidopsis exocyst subunits possess consensus Atg8-interacting motifs (AIMs), which may mediate their interaction with the autophagy-associated Atg8 protein, providing thus a mechanistic base for participation of exocyst (sub)complexes in autophagy. However, the bioinformatically identified AIMs are short peptide motifs that may occur by chance. We thus performed an exhaustive search in a large collection of plant exocyst-derived sequences from our previous bioinformatic study and found that AIMs are over-represented among exocyst subunits of all lineages examined, including moss and club moss, compared with a representative sample of the Arabidopsis proteome. This is consistent with the proposed exocyst AIMs being biologically meaningful and evolutionarily ancient. Moreover, among the numerous EXO70 paralogs, the monocot-specific EXO70F clade appears to be exempt from the general AIM enrichment, suggesting a modification of the autophagy connection in a subset of exocyst variants.
The hetero-octameric exocyst complex, historically associated mainly with Golgi to plasmalemma membrane trafficking,Citation1-Citation3 or at least its subunits and/or subcomplexes, was recently found to participate in autophagy in both metazoansCitation4 and plants. EXO70B1, one of the 23 EXO70 paralogs of Arabidopsis thaliana, is required for autophagy and vacuole biogenesis, colocalizes with the Atg8f autophagic marker, and probably participates, together with EXO84B and SEC5A, in an “autophagic” exocyst sub-complex.Citation5 Bioinformatic analysis of 20 out of the 35 exocyst subunits encoded by the A. thaliana genome revealed numerous consensus Atg8-interacting motifs (AIMs),Citation6 indicating a possible direct interaction between exocyst subunits and the Atg8 family of autophagy regulators.Citation7
However, AIMs are short peptide motifs with a degenerate consensus – X(3)-[FYW]-X(2)-[VIL]-X(3) where the X positions are enriched in negatively charged amino acids (D,E) – that may pick false positives in pattern searches, and the presence of a consensus sequence alone may not be sufficient to conclude that the pattern is biologically relevant, i.e., that it binds the Atg8 protein in vivo (albeit its evolutionary conservation, as reported for one AIM within EXO70A1 and B1,Citation6 might be considered as additional evidence).
We thus decided to test whether exocyst subunits contain more AIMs than a randomly selected representative sample of plant protein sequences. To estimate a “baseline” distribution of AIMs, we employed the public PatMatchCitation8 server at The Arabidopsis Information Resource (TAIR, http://www.Arabidopsis.org) to search the Arabidopsis proteome (TAIR10 reference protein sequences) for all versions of the AIM patterns from the recently published report.Citation6 From the results, a non-redundant list of AIMs present in the 7078 predicted proteins from A. thaliana Chromosome I was generated using built-in functions of Microsoft Excel, and distribution of AIM hit numbers per protein was estimated. Only very few protein sequences contained more than 2 AIMs ( and ). Then, standalone PatMatch (downloaded from the TAIR website) was used to search for AIMs in 382 exocyst subunit sequences assembled in our previous study on exocyst evolution,Citation9 covering all subunits from a variety of plant lineages including Physcomitrella, Selaginella, and several monocot (grasses) and dicot angiosperms. Exocyst subunits contained, in general, dramatically more AIMs than the baseline proteins. All lineages examined (including moss and club moss representatives) behaved similarly, except the grasses, where the enrichment was far less pronounced (). The distribution of AIM frequencies in non-grass exocyst subunits appears to be lineage-independent, justifying the use of baseline data from Arabidopsis at least for non-grass lineages. The EXO70 subunits alone exhibited AIMs distribution similar to that found for the whole exocyst (). Since the numerous EXO70 paralogs form the majority of our exocyst sequence collection, they are the main source of the different AIM distribution in grasses.
Figure 1. (A) AIM frequency profiles of exocyst subunits from several plant lineages. Number of AIM hits per protein on the abscissa (x), fraction of the total protein set analyzed on the ordinate (y). (B) AIM frequency profiles for the EXO70 subunits.
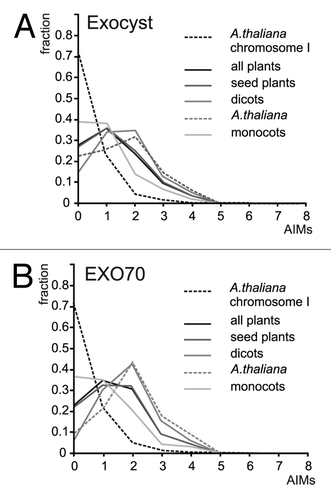
Figure 2. (A) A schematic phylogenetic tree of the EXO70 subunits, showing the major clades (based on Figure 3 from ref. Citation9). “BNG” is a “basal non-angiosperm group” consisting of moss and lycophyte sequences. (B) AIM frequency profiles for EXO70.1 and EXO70.3 clades. (C) AIM frequency profiles for the EXO70.2 clade with exclusion of the F and FX branches. Note that EXO70C and H (top graph) contain generally fewer AIMs thanEXO70B, D and F (bottom graph). (D) AIM frequency profiles for the EXO70F branch showing that the low frequency of AIMs in this clade can be attributed to the grass-specific FX group.
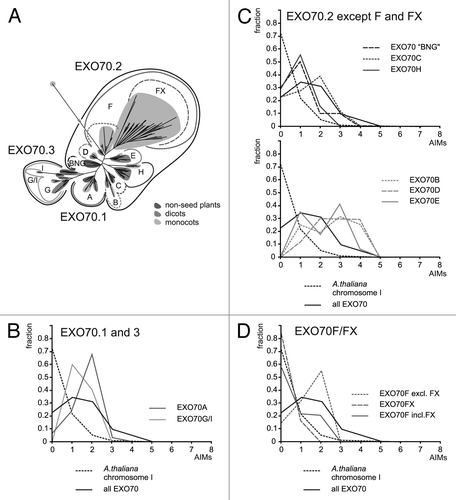
In our recent study of Exocyst evolutionCitation9 we confirmed the results of previous analyses suggesting that the diversification of EXO70s started already in the common ancestor of land plants, which had 3 EXO70 lineages ().Citation10-Citation12 One of them, EXO70.2, underwent subsequent extensive radiation that generated 6 major clades in angiosperms (EXO70 B,C,D,E,F, and H). Among these, EXO70H considerably expanded in the dicots, and its member EXO70H1 was shown to participate in pathogen defense in Arabidopsis together with EXO70B2, a close relative of the “autophagic” EXO70B1,Citation13 raising thus the possibility that the enormous diversity of the dicot EXO70H clade may be linked to biotic stress responses. In the monocots EXO70H radiation did not take place, but a novel major branch (termed EXO70FX), which may be grass-specific, evolved within the EXO70F clade.Citation9
An examination of the AIM frequency profiles in distinct EXO70 clades revealed that all of them except F are AIM-enriched to a lesser or greater extent, with EXO70B, D, and E containing most motifs ( and C). Within the F clade, the FX branch does not noticeably differ from the baseline (Chromosome I) profile, while “classical” EXO70Fs (including those from grasses) follow the overall exocyst pattern (). Thus, the difference between the monocots/grasses and the remaining lineages examined is not due to some overall feature of the grass or monocot proteomes or their EXO70s, but rather appears to be a specific consequence of the radiation of EXO70FXs, which apparently have lost the ancestral AIM enrichment. We have previously reported that most members of all 3 major EXO70 groups, with exception of EXO70FX, harbor up to 2 conserved peptide motifs at their otherwise variable N-termini.Citation9 Remarkably, one of these motifs (motif 2) in most proteins from the EXO70.1 and EXO70.3 branches contains the standard AIM pattern with consensus [DE](2)-E-F-X(2)-[IL]-X(3), often with additional D or E at the X positions (see Figure 4 in ref. Citation9); this also is the motif recognized as common to EXO70A1 and EXO70B1 previously.Citation6
The distribution of AIM frequencies in exocyst proteins, together with recent experimental observations from both plants and metazoans, suggests that involvement of the exocyst complex in autophagy is evolutionarily conserved, and that a subset of EXO70 paralogs in (at least some) monocots has been modified in respect to their autophagic roles. This then relieved the selection pressure maintaining the interface(s) for binding the Atg8 protein, or even led to selective elimination of the AIMs to keep EXO70FX protected against autophagy in the context of grass pathogen defense.
To test if the presence of AIMs, which themselves are evolutionarily conserved from yeast to mammals,Citation14 is an ancestral feature of the exocyst subunits, we performed additional PatMatch searches on 17 exocyst subunit sequences from budding yeast and mouse, found by keyword searches of the reference section of GenBank. Consistent with Atg8 binding being common in exocyst proteins, all subunits except mouse Sec10, mouse Exo84, and yeast Sec8 contained 1 to 3 AIMs (however, the plant-derived AIM consensus might have been too restrictive for opisthokont sequence searches).
The autophagic pathway may thus be on the way toward joining other ancient features of eukaryotic cells that possibly date back to the last eukaryotic common ancestor (LECA), such as, e.g., the central cell cycle regulation machinery,Citation15 or the complex apparatus ensuring the cytoskeletal rearrangements and membrane-trafficking processes required for cell invasivity.Citation16
| Abbreviations: | ||
| AIM | = | Atg8 interacting motif |
| LECA | = | last eukaryotic common ancestor |
| EXO70 | = | exocyst complex subunit 70 |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work has been supported by the GACR P305/11/1629 grant from the Grant Agency of the Czech Republic and by the EU PLANTORIGINS-ITN 238640 project.
References
- He B, Guo W. The exocyst complex in polarized exocytosis. Curr Opin Cell Biol 2009; 21:537 - 42; http://dx.doi.org/10.1016/j.ceb.2009.04.007; PMID: 19473826
- Zhang Y, Liu CM, Emons AM, Ketelaar T. The plant exocyst. J Integr Plant Biol 2010; 52:138 - 46; http://dx.doi.org/10.1111/j.1744-7909.2010.00929.x; PMID: 20377676
- Heider MR, Munson M. Exorcising the exocyst complex. Traffic 2012; 13:898 - 907; http://dx.doi.org/10.1111/j.1600-0854.2012.01353.x; PMID: 22420621
- Bodemann BO, Orvedahl A, Cheng T, Ram RR, Ou YH, Formstecher E, Maiti M, Hazelett CC, Wauson EM, Balakireva M, et al. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell 2011; 144:253 - 67; http://dx.doi.org/10.1016/j.cell.2010.12.018; PMID: 21241894
- Kulich I, Pečenková T, Sekereš J, Smetana O, Fendrych M, Foissner I, Höftberger M, Žárský V. Arabidopsis exocyst subcomplex containing subunit EXO70B1 is involved in autophagy-related transport to the vacuole. Traffic 2013; 14:1155 - 65; PMID: 23944713
- Tzfadia O, Galili G. The Arabidopsis exocyst subcomplex subunits involved in a golgi-independent transport into the vacuole possess consensus autophagy-associated atg8 interacting motifs. Plant Signal Behav 2013; 9:e26732; http://dx.doi.org/10.4161/psb.26732; PMID: 24168876
- Honig A, Avin-Wittenberg T, Ufaz S, Galili G. A new type of compartment, defined by plant-specific Atg8-interacting proteins, is induced upon exposure of Arabidopsis plants to carbon starvation. Plant Cell 2012; 24:288 - 303; http://dx.doi.org/10.1105/tpc.111.093112; PMID: 22253227
- Yan T, Yoo D, Berardini TZ, Mueller LA, Weems DC, Weng S, Cherry JM, Rhee SY. PatMatch: a program for finding patterns in peptide and nucleotide sequences. Nucleic Acids Res 2005; 33:W262-6; http://dx.doi.org/10.1093/nar/gki368; PMID: 15980466
- Cvrčková F, Grunt M, Bezvoda R, Hála M, Kulich I, Rawat A, Žárský V. Evolution of the land plant exocyst complexes. Front Plant Sci 2012; 3:159; http://dx.doi.org/10.3389/fpls.2012.00159; PMID: 22826714
- Eliáš M, Drdová E, Žiak D, Bavlnka B, Hála M, Cvrčková F, Soukupová H, Žárský V. The exocyst complex in plants. Cell Biol Int 2003; 27:199 - 201; http://dx.doi.org/10.1016/S1065-6995(02)00349-9; PMID: 12681307
- Synek L, Schlager N, Eliás M, Quentin M, Hauser MT, Žárský V. AtEXO70A1, a member of a family of putative exocyst subunits specifically expanded in land plants, is important for polar growth and plant development. Plant J 2006; 48:54 - 72; http://dx.doi.org/10.1111/j.1365-313X.2006.02854.x; PMID: 16942608
- Chong YT, Gidda SK, Sanford C, Parkinson J, Mullen RT, Goring DR. Characterization of the Arabidopsis thaliana exocyst complex gene families by phylogenetic, expression profiling, and subcellular localization studies. New Phytol 2010; 185:401 - 19; http://dx.doi.org/10.1111/j.1469-8137.2009.03070.x; PMID: 19895414
- Pečenková T, Hála M, Kulich I, Kocourková D, Drdová E, Fendrych M, Toupalová H, Žárský V. The role for the exocyst complex subunits Exo70B2 and Exo70H1 in the plant-pathogen interaction. J Exp Bot 2011; 62:2107 - 16; http://dx.doi.org/10.1093/jxb/erq402; PMID: 21199889
- Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett 2010; 584:1379 - 85; http://dx.doi.org/10.1016/j.febslet.2010.01.018; PMID: 20083108
- Guo Z, Stiller JW. Comparative genomics of cyclin-dependent kinases suggest co-evolution of the RNAP II C-terminal domain and CTD-directed CDKs. BMC Genomics 2004; 5:69; http://dx.doi.org/10.1186/1471-2164-5-69; PMID: 15380029
- Vaškovičová K, Žárský V, Rösel D, Nikolič M, Buccione R, Cvrčková F, Brábek J. Invasive cells in animals and plants: searching for LECA machineries in later eukaryotic life. Biol Direct 2013; 8:8; http://dx.doi.org/10.1186/1745-6150-8-8; PMID: 23557484