Abstract
In aerobes, anoxia impairs mitochondrial energy generation as well as biosynthesis and degradation of essential cell components. In a recent analysis we have shown that the unicellular green alga Chlamydomonas reinhardtii responds to anaerobiosis in the dark by significant changes of the transcriptome, which, in summary, were directed at saving and economizing energy. Several of the transcriptional changes were related to photosynthesis and were accompanied by reduced amounts of chlorophylls and plastid lipids as well as lowered photosystem 2 quantum yields. A further noticeable pattern was a transcriptional upregulation of various genes encoding O2 dependent enzymes of central biosynthetic pathways. However, cells do not divide in dark-anoxia, indicating that C. reinhardtii cannot compensate for the lack of O2 and light. Upon return to aeration and light, cultures show severe photo-bleaching, which might be a stress reaction, but also part of an acclimation process or its disturbance.
Although they generate molecular oxygen (O2) during oxygenic photosynthesis, many green organisms can find themselves confronted with hypoxic or anoxic conditions in shaded aquatic environments or in waterlogged soil. O2 deficiency has a profound influence on the energy metabolism, and strategies of hypoxia-tolerant species like rice to cope with low O2 concentrations involve quiescence strategies and the economic utilization of carbohydrates,Citation1 showing the importance of the energy status for surviving longer periods of anaerobiosis.
In a recent study, we analyzed the responses of the unicellular green alga Chlamydomonas reinhardtii to dark-anoxia using an RNA-Seq approach.Citation2 The alga has an elaborate and flexible ?bacterial-type? fermentation metabolism,Citation3,Citation4 and the wild type grows without any difficulty in an anaerobic atmosphere in the light.Citation2,Citation5-Citation7 However, what could be deduced from transcript patterns and accompanying physiological analyses of cells subjected to anoxia in darkness was in accordance with a transition to a ?power save mode?: The cells stopped dividing and exhibited an overall decrease of the amounts of transcripts associated with growth and gene expression. A significant amount of upregulated transcripts related to starch, amino acid, and fatty acid catabolism furthermore indicated that the microalgae sought to make all internal energy sources available.
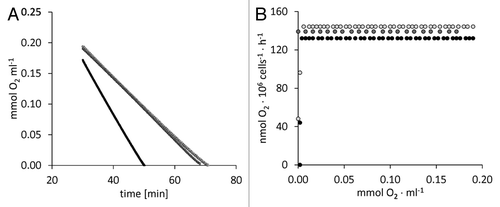
However, not only the energy metabolism is impaired when O2 is limiting. The biosynthesis and degradation of various cell compounds such as fatty acids or tetrapyrroles requires O2 as a substrate or reagent. Though under debate, it has been discussed that upon O2 limitation, plants might actively downregulate the rate of respiratory O2 uptake in order to maintain a certain O2 threshold for essential metabolic reactions.Citation8 For the transcriptome study, C. reinhardtii was simply transferred to gas-tight flasks in the dark, allowing the cells to ?anaerobize themselves? by respiratory activity. We analyzed if C. reinhardtii would consume dissolved O2 completely when placed into a sealed container (). Indeed, the cells kept the respiratory rate unchanged until O2 became undetectable (), indicating that no or only minute amounts of O2 are maintained within the cells.
Figure 1. Three liquid cultures of C. reinhardtii strain CC-124 were grown in Tris Acetate Phosphate (TAP) medium on a horizontal shaker at 20 °C and with bottom-up illumination of 80 µmol photons ? m?2 ? s?1 up to a cell density of 2.5 (light gray symbols), 2.7 (dark gray symbols), or 3.5 (black symbols) ? 106 cells ml?1. 2-ml aliquots of the cell suspensions were transferred to the sealed measuring cell of a Hansatech O2 electrode according to Philipps et al., 2011.Citation19 O2 uptake was monitored in darkness until O2 became undetectable (A). (B) O2 uptake rates were calculated each minute and plotted against the respective O2 concentration.
The generation of complete anoxia notwithstanding, the RNA-Seq data of dark-anoxic C. reinhardtii cells revealed an accumulation of various transcripts encoding O2 dependent enzymes. Among those were several fatty acid desaturases, enzymes of tetrapyrrole and sterol biosynthesis as well as prolyl-4-hydroxylases. The desaturation status of the fatty acids from dark-anoxic algal cells, however, was not enhanced,Citation2 and we concluded that the upregulation of transcripts encoding O2-requiring enzymes is probably a?futile?compensating response. Spinning this thought further, O2-dependent enzymatic reactions or their products might well be used as O2 sensors in C. reinhardtii. To date, the COPPER RESPONSE REGULATOR1 (CRR1) is the only specific transcription factor that is known to regulate a subset of genes in response to anoxia in Chlamydomonas.Citation7,Citation9-Citation11 Comparing transcriptome profiles of O2 deprived crr1 mutants with those of the rescued strain (crr1:CRR1) confirmed the well-known connection between copper and O2 deficiency mediated by CRR1. However, the differential expression of only about 40 transcripts depended on CRR1,Citation2 showing that further O2 sensing and -signaling strategies must exist to account for the major part of the transcriptional changes. In yeast, products of O2-requiring biosynthetic pathways, heme and sterols, are involved in O2 sensing, while in mammals, O2-dependent hydroxylation by prolyl-4-hydroxylases of the central transcription factor HIF1? controls its proteasomal turnover.Citation12,Citation13 Arabidopsis group VII Ethylene Response Factors (ERFs), transcription factors that regulate low oxygen responses, are destabilized in normoxia by the N-end rule: The N-terminal Cys residue becomes oxidized under normoxic conditions, possibly by an O2-requiring enzymatic reaction.Citation14 In parallel to these examples, and in view of the upregulation of genes coding for O2-dependent enzymes, one might speculate that C. reinhardtii makes a virtue out of necessity and utilizes the inability to carry out these reactions as a means of O2 sensing.
The dependence of (metabolic and/or regulative) enzymes on the O2 molecule can also result in the inability to catalyze O2-dependent degradation steps, and this might be a further challenge. Tetrapyrroles are good representatives, because these molecules as well as some pre-cursors and degradation intermediates are photosensitizers.Citation15,Citation16 For example, an impairment of the O2-requiring pheophorbide a oxygenase, involved in chlorophyll (Chl) breakdown, might result in the accumulation of pheophorbide a, which is a photosensitizer, but also involved in programmed cell death.Citation17 The dark-anoxic C. reinhardtii strains analyzed by RNA-Seq had lower Chl contents and exhibited decreased abundances of transcripts encoding tetrapyrrole biosynthetic enzymes and Chl-binding proteins.Citation2 This was accompanied by a moderate upregulation of genes encoding a NON YELLOW COLORING1 (NYC1) homolog and a putative chlorophyllase (CLH1). NYC1 is a short-chain dehydrogenase likely to be a Chl b reductase and, together with NYC1-LIKE (NOL), important for Chl breakdown during senescence,Citation15,Citation17 while chlorophyllase removes the phytol tail from Chl and, besides pheophytinase, might also be involved in Chl degradation.Citation15
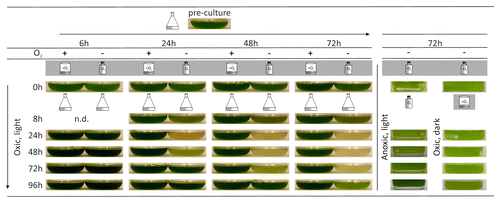
Because the transcriptome study indicated that dark-anoxic incubation had a significant influence on the Chl metabolism of C. reinhardtii,Citation2 possibly resulting in an accumulation of Chl degradation intermediates, a photo-bleaching phenotype after oxic or anoxic incubation in the dark for increasing periods of time (up to 72 h) was analyzed. Cells incubated in sealed flasks, i.e., anoxically, for 6 h did not show any chlorosis when transferred back to aerated and illuminated conditions (). However, after 24 to 72 h of incubation, severe photo-bleaching was observed, and the recovery back to green cultures took more time the longer the cells had been subjected to dark-anoxia (). The fact that the bleaching of the cells was dependent on light and normal aeration became apparent by re-oxygenating cells in darkness or re-illuminating them in sealed flasks. Cells such treated did not show signs of chlorosis ().
Figure 2.C. reinhardtii strain CC-124 cultures were grown as described in the legend of up to a cell density of about 3 ? 106 cells ml?1. Then they were transferred to darkness either aerated in beakers (O2 +) or in sealed flasks (O2 ?). Cells were incubated under these conditions for 6, 24, 48, or 72 h (indicated in the top row) and then transferred back to standard growth conditions (aeration and illumination at 80 µmol photons ? m?2 ? s?1; Oxic, light). Aliquots of cells incubated dark-anoxically for 72 h were also kept in sealed flasks, but re-illuminated (Anoxic, light) or re-oxygenated in the dark (Oxic, dark). Photographs were taken immediately after transfer of the cells to Erlenmeyer flasks (0h at the left) and then after 8, 24, 48, 72, and 96 h (indicated at the left).
Whether the observed photo-bleaching of C. reinhardtii is specific for a dark-anoxic influence on Chl (or tetrapyrrole) biosynthesis, -binding and -degradation remains to be examined. A potentially stressful consequence of anoxia and re-aeration is a (photosensitizer-independent) generation of reactive oxygen and nitrogen species (ROS, RNS), which are produced and employed as second messengers by plants under anoxia.Citation18 Various transcripts related to ROS and RNS generation and -signaling indicated that C. reinhardtii shares the ubiquitously observed anoxic generation and utilization of ROS and RNS,Citation2 and we could show that NO is involved in the anoxic response of C. reinhardtii.Citation5 We also noted transcript patterns speaking for an activation of ROS/RNS stress defense systems. Sudden re-aeration in combination with light might overstrain the antioxidative defense systems of C. reinhardtii cells. However, the photo-bleaching phenotype might also be part of a long-term acclimatory process to anoxia in darkness. The RNA-Seq profiles revealed many genes involved in photosynthesis and Chl binding to be downregulated, and though this transcriptional response was probably due to darkness rather than O2 deficiency, only dark-anoxic cells exhibited strongly decreased photosystem 2 quantum yields and lowered amounts of the plastid lipids monogalactosyl- and digalactosyldiacylglycerol (MGDG and DGDG).Citation2 Furthermore, the decreased amounts of MGDG and DGDG were accompanied by the appearance of typical MGDG- and DGDG-associated unsaturated fatty acids in triacylglycerols. Though speculative for the time being, it might be proposed that dark-anoxic C. reinhardtii cells specifically dismantle their photosynthetic apparatus, either to reallocate resources or to be prepared for sudden and potentially harmful re-oxygenation and -illumination. As we additionally observed significant effects of darkness and of inhibiting photosystem 2 with 3-(3,4-Dichlorophenyl)-1,1-dimethylurea (DCMU) on transcript abundances of ?anaerobic genes?Citation2, the metabolic and regulatory interactions of the photosynthetic machinery and (dark-) anoxic responses appear to be an important theme in C. reinhardtii.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG), (He 5790/1?1 and -2, and HE 5790/3?1).
References
- Bailey-Serres J, Voesenek LA. Flooding stress: acclimations and genetic diversity. Annu Rev Plant Biol 2008; 59:313 - 39; http://dx.doi.org/10.1146/annurev.arplant.59.032607.092752; PMID: 18444902
- Hemschemeier A, Casero D, Liu B, Benning C, Pellegrini M, Happe T, Merchant SS. COPPER RESPONSE REGULATOR1-Dependent and -Independent Responses of the Chlamydomonas reinhardtii Transcriptome to Dark Anoxia. Plant Cell 2013; 25:3186 - 211; http://dx.doi.org/10.1105/tpc.113.115741; PMID: 24014546
- Catalanotti C, Yang W, Posewitz MC, Grossman AR. Fermentation metabolism and its evolution in algae. Front Plant Sci 2013; 4:150; http://dx.doi.org/10.3389/fpls.2013.00150; PMID: 23734158
- Hemschemeier A, Happe T. Alternative photosynthetic electron transport pathways during anaerobiosis in the green alga Chlamydomonas reinhardtii. Biochim Biophys Acta 2011; 1807:919 - 26; http://dx.doi.org/10.1016/j.bbabio.2011.02.010; PMID: 21376011
- Hemschemeier A, Düner M, Casero D, Merchant SS, Winkler M, Happe T. Hypoxic survival requires a 2-on-2 hemoglobin in a process involving nitric oxide. Proc Natl Acad Sci U S A 2013; 110:10854 - 9; http://dx.doi.org/10.1073/pnas.1302592110; PMID: 23754374
- Eriksson M, Moseley JL, Tottey S, Del Campo JA, Quinn J, Kim Y, Merchant S. Genetic dissection of nutritional copper signaling in chlamydomonas distinguishes regulatory and target genes. Genetics 2004; 168:795 - 807; http://dx.doi.org/10.1534/genetics.104.030460; PMID: 15514054
- Quinn JM, Eriksson M, Moseley JL, Merchant S. Oxygen deficiency responsive gene expression in Chlamydomonas reinhardtii through a copper-sensing signal transduction pathway. Plant Physiol 2002; 128:463 - 71; http://dx.doi.org/10.1104/pp.010694; PMID: 11842150
- Gupta KJ, Zabalza A, van Dongen JT. Regulation of respiration when the oxygen availability changes. Physiol Plant 2009; 137:383 - 91; http://dx.doi.org/10.1111/j.1399-3054.2009.01253.x; PMID: 19549068
- Quinn JM, Barraco P, Eriksson M, Merchant S. Coordinate copper- and oxygen-responsive Cyc6 and Cpx1 expression in Chlamydomonas is mediated by the same element. J Biol Chem 2000; 275:6080 - 9; http://dx.doi.org/10.1074/jbc.275.9.6080; PMID: 10692397
- Kropat J, Tottey S, Birkenbihl RP, Depège N, Huijser P, Merchant S. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc Natl Acad Sci U S A 2005; 102:18730 - 5; http://dx.doi.org/10.1073/pnas.0507693102; PMID: 16352720
- Castruita M, Casero D, Karpowicz SJ, Kropat J, Vieler A, Hsieh SI, Yan W, Cokus S, Loo JA, Benning C, et al. Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 2011; 23:1273 - 92; http://dx.doi.org/10.1105/tpc.111.084400; PMID: 21498682
- Bailey-Serres J, Chang R. Sensing and signalling in response to oxygen deprivation in plants and other organisms. Ann Bot 2005; 96:507 - 18; http://dx.doi.org/10.1093/aob/mci206; PMID: 16051633
- Bien CM, Espenshade PJ. Sterol regulatory element binding proteins in fungi: hypoxic transcription factors linked to pathogenesis. Eukaryot Cell 2010; 9:352 - 9; http://dx.doi.org/10.1128/EC.00358-09; PMID: 20118213
- Bailey-Serres J, Fukao T, Gibbs DJ, Holdsworth MJ, Lee SC, Licausi F, Perata P, Voesenek LA, van Dongen JT. Making sense of low oxygen sensing. Trends Plant Sci 2012; 17:129 - 38; http://dx.doi.org/10.1016/j.tplants.2011.12.004; PMID: 22280796
- Tanaka R, Kobayashi K, Masuda T. Tetrapyrrole Metabolism in Arabidopsis thaliana.. Arabidopsis Book 2011; 9:e0145; http://dx.doi.org/10.1199/tab.0145; PMID: 22303270
- Mochizuki N, Tanaka R, Grimm B, Masuda T, Moulin M, Smith AG, Tanaka A, Terry MJ. The cell biology of tetrapyrroles: a life and death struggle. Trends Plant Sci 2010; 15:488 - 98; http://dx.doi.org/10.1016/j.tplants.2010.05.012; PMID: 20598625
- Hörtensteiner S, Kräutler B. Chlorophyll breakdown in higher plants. Biochim Biophys Acta 2011; 1807:977 - 88; http://dx.doi.org/10.1016/j.bbabio.2010.12.007; PMID: 21167811
- Blokhina O, Fagerstedt KV. Oxidative metabolism, ROS and NO under oxygen deprivation. Plant Physiol Biochem 2010; 48:359 - 73; http://dx.doi.org/10.1016/j.plaphy.2010.01.007; PMID: 20303775
- Philipps G, Happe T, Hemschemeier A. Nitrogen deprivation results in photosynthetic hydrogen production in Chlamydomonas reinhardtii.. Planta 2012; 235:729 - 45; http://dx.doi.org/10.1007/s00425-011-1537-2; PMID: 22020754