Abstract
Jasmonates have crucial roles in plant responses to biotic and abiotic stresses. Given the importance of transcriptional regulation in jasmonate-mediated stress responses, transcription factors are key regulators of jasmonate signaling. The transcription factors JASMONATE-ASSOCIATED MYC2-LIKE 1 (JAM1), JAM2, and JAM3 are negative regulators of jasmonate signaling, although the mechanisms that control the activities of these transcription factors remain unclear. To understand the regulatory mechanisms of JAM proteins, we used a yeast two-hybrid assay to screen for protein interaction partners of JAM1 and found that JAM1 interacted with JAZ proteins.
As JAZ proteins interact with MYC2 and suppresses its transcriptional activity, we performed comprehensive interaction analysis between JAM1 through JAM3 and JAZ1 through JAZ12. We showed that JAM1 and JAM2 interacted with several of the JAZ proteins analyzed here. In contrast, JAM3 interacted with none of the tested JAZ proteins. These results suggest that JAZ proteins regulate JAM1 and JAM2 through protein-protein interactions in a manner similar to the repressive effects of JAZ proteins on the transcriptional activity of MYC2.
Introduction
Jasmonic acid (JA) and its cyclic precursors and derivatives, the so-called jasmonates, are growth and stress response regulators that are ubiquitous throughout the plant kingdom. Jasmonates are rapidly synthesized in response to certain external stimuli, such as wounding, pathogen infection, and insect attack.Citation1-Citation3 Those stresses and JA treatment each induce similar sets of jasmonate-responsive genes, which are involved in defense-related metabolism.Citation4-Citation9
Jasmonoyl-l-isoleucine (JA-Ile), which is an active form of jasmonate,Citation10,Citation11 promotes interaction between COI1 and JAZ proteins, and then JAZ proteins are presumably degraded via the 26S proteasome system.Citation12-Citation14 MYC2, as well as its close homologs MYC3 and MYC4, is a central transcriptional regulator of jasmonate signaling and is negatively regulated by JAZ proteins through protein interactions. The degradation of JAZ proteins results in de-repression of the MYC transcription factors, which leads to transcriptional activation of downstream jasmonate-responsive genes.Citation15-Citation20 The N-terminal regions of MYC2 and MYC3 each interact with JAZ proteins.Citation18 Moreover, the formation of homo- and heterodimers by the MYC transcription factorsCitation18 suggests the importance of protein interactions among those factors that are required for jasmonate signaling.
JA-ASSOCIATED MYC2-LIKE 1 (JAM1), JAM2 and JAM3, which are homologs of MYC2, were recently shown to function redundantly as negative regulators of jasmonate signaling because the jam1jam3jam3 triple mutants (jam×3) shows enhanced responsiveness to jasmonate, JAM1, JAM2 and JAM3 have repression activities, and they form homo-and hetero dimers.Citation21-Citation24 Here, we screened for interaction partners of JAM1 and further showed that JAM1 and JAM2, but not JAM3, interacted with several JAZ proteins. We concluded that JAZ proteins are possible regulators of JAM transcription factors.
Results
Identification of interaction targets of JAM transcription factors
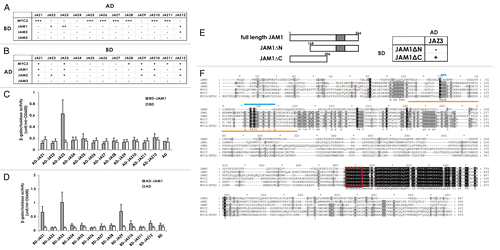
Yeast two-hybrid screens using JAM1 as bait identified 12 positive clones, which were sequenced to identify JAM1 targets (data not shown). Ten of the 12 were JAZ proteins such as JAZ1 (At1g19180, 2 clones), JAZ2 (At1g74950), JAZ3 (At3g17860, 4 clones), JAZ9 (At1g70700), JAZ10 (At5g13220) and JAZ12 (At5g20900), and the remaining two clones were JAM3 and HY1 (heme oxygenase, At2g26670). To test whether JAM1 interacts specifically with other JAZ proteins, we used yeast two-hybrid assays to check all possible combinations between full-length bHLH transcription factors (BD-JAM1 through BD-JAM3 and BD-MYC2 as bait) and full-length AD-JAZ1 through AD-JAZ12 proteins (as prey). We also tested the opposite combination (BD-JAZ proteins as bait and AD-bHLH proteins as prey) (). JAM1 interacted with JAZ3 and JAZ12, and JAM2 interacted with JAZ12 in both sets of experiments. JAM3 did not interact with any JAZ proteins under the experimental conditions used. As previously reported, MYC2 interacted with several JAZ proteins, with the exception of JAZ4 ().Citation18,Citation25 To quantify the possible interactions between JAM1 and the 12 JAZ proteins, we measured the β-galactosidase activity of cell cultures that expressed those proteins. Detection of β-galactosidase activity in cell cultures that expressed the combinations of BD-JAM1 and AD-JAZ3 () and BD-JAZ1, -JAZ3, or -JAZ9 and AD-JAM1 () confirmed the interactions of these proteins in yeast. We also tested the interaction between truncated derivatives of JAM1 and full-length JAZ3. The interaction of JAM1ΔC, but not JAM1ΔN, with JAZ3 indicated the necessity of the N-terminal region for the interaction of JAM1 with JAZ3 (), which is also true for the interaction between N-terminal region of JAZ3 and MYC2 or MYC3.Citation18
Figure 1. A. Protein-protein interaction between bHLH transcription factors and JAZ proteins. A summary of yeast two-hybrid results to assay for interactions between bHLH transcription factors as bait (BD) and JAZ repressors as prey (AD) (A), and the opposite combination (JAZ proteins as bait and bHLH proteins as prey) (B). AD, DNA activation domain; BD, DNA binding domain. Based on the number of colonies formed, the strength of each interaction was rated as strong (+++), medium (++), weak (+), or undetectable (–). (C), (D) Quantitative assays of β-galactosidase activity using o-nitrophenyl-β-d-galactopyranose as a substrate to determine protein interactions between JAM1 and JAZ proteins. Results are shown as the mean ± SD for biologically independent experiments (n = 3). (E) The interaction of truncated JAM1 derivatives with full-length JAZ3 was tested. (F) Alignment of the amino acid sequences of JAM1, JAM2, JAM3, MYC2, MYC3, and MYC4. Identical and similar amino acids are shaded black and gray, respectively. Red boxes indicate the nuclear localization signals. Orange lines under the sequences indicate the JAZ interaction domain of MYC2 and MYC3.Citation18 Light blue lines above the sequences indicate the conserved amino acid residues within a JAZ-interacting domain among JAM1, JAM2, MYC2, MYC3, and MYC4, but not JAM3. . Protein-protein interaction between bHLH transcription factors and JAZ proteins. A summary of yeast two-hybrid results to assay for interactions between bHLH transcription factors as bait (BD) and JAZ repressors as prey (AD) (E) The interaction of truncated JAM1 derivatives with full-length JAZ3 was tested. (F) Alignment of the amino acid sequences of JAM1, JAM2, JAM3, MYC2, MYC3, and MYC4. Identical and similar amino acids are shaded black and gray, respectively. Red boxes indicate the nuclear localization signals. Orange lines under the sequences indicate the JAZ interaction domain of MYC2 and MYC3.Citation18 Light blue lines above the sequences indicate the conserved amino acid residues within a JAZ-interacting domain among JAM1, JAM2, MYC2, MYC3, and MYC4, but not JAM3.
Expression analysis of JAZ genes in jam×3 plants
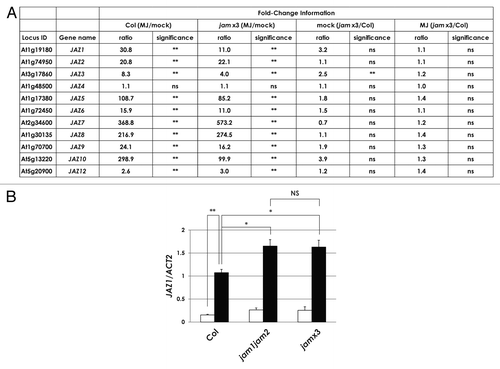
Given that JAM proteins negatively regulate the expression of jasmonate-responsive genes,Citation21-Citation23 we focused on the expression level of JAZ genes in jam×3 plants, which are loss of function mutant of JAM1, JAM2 and JAM3.Citation21 shows a summary of expression levels of JAZ genes obtained from GeneChip analyses.Citation21 The expression of all JAZ genes, except JAZ4, was induced by MJ treatment both in wild-type plants of the ecotype Columbia (Col) and in jam×3 plants. With the exception of JAZ3, the expression levels of all JAZ genes in mock-treated Col and jam×3 plants were comparable as well as in MJ treated plants (). Thus the expression levels of JAZ genes are almost comparable between Col and jam×3 in spite of the presence or absence of MJ. We further tested the expression levels of JAZ1 in Col, jam1jam2, and jam×3 plants by qRT-PCR analysis in more detail (). The expression level of JAZ1 in jam1jam2 and jam×3 plants was only 1.5-fold higher than that in Col plants (P < 0.05). The almost comparable expression levels of JAZ1 in jam1jam2 and jam×3 plants indicated that the negative effect on JAZ1 expression derives from JAM1 and JAM2 but such effect is limited.
Figure 2. Expression analysis of JAZ genes in jam×3 plants. (A) A summary of fold-change information and statistical analyses of JAZ expression levels, based on the normalized intensity of JAZ genes obtained from GeneChip analyses.Citation21 Results are shown as the mean for two biologically independent RNA samples. ** P < 0.01, * P < 0.05, NS = not significant (P ≥ 0.05); Tukey-Kramer multiple comparison test. (B) Total RNA was isolated from 7-d-old plants, which were grown in GM liquid medium with 1% sucrose and treated with mock (white bars) or 50 μM MJ (black bars) for 1 h. Results are shown as the mean ± SE for three biologically independent RNA samples.
Discussion
Our yeast two-hybrid analysis indicated that JAM1 and JAM2 interact with JAZ proteins (). The interaction of the N-terminal region of JAM1 with JAZ3 () indicated the existence of a JAZ-interaction motif in that region. The N-terminal region of MYC2 (Y93 to E160) is necessary for the interaction of MYC2 with JAZ proteins (, orange lines).Citation18 This region contains several amino acids that are conserved in MYC2 through MYC4, as well as in JAM1 and JAM2, but not in JAM3 (, pale blue lines). Given that JAM3 did not interact with any JAZ proteins in our experimental conditions, these results imply that these amino acids are involved in the interaction of JAZ proteins and JAM proteins.
JAZ proteins interact with bHLH transcription factors via a C-terminal region, which contains a Jas motif,Citation22,Citation25 and also interact with COI1.Citation26,Citation27 JA-Ile–dependent formation of protein complexes that contain COI1 and JAZ proteins is followed by the degradation of JAZ proteins. Thus, removal of the COI1-intereacting region of JAZ proteins might stabilize them and thereby compromise the expression of jasmonate-responsive genes and suppress jasmonate responses by blocking the transcriptional activity of transcription factors, such as MYC2.Citation12,Citation13 Our demonstration here that some JAZ proteins interacted with JAM1 and JAM2 () led us to speculate that physical interactions between JAZ and JAM proteins might inhibit the function of JAM proteins as transcriptional repressor. It is not clear whether the JAM and JAZ proteins could co-localize in the same cell. Therefore, further research is required to determine the protein interaction among those proteins in vivo. The expression of JAZ genes is induced by MYC2, MYC3, and MYC4,Citation12,Citation18 and the expression of JAM genes is partially MYC2-dependent.Citation21 Although JAM proteins negatively regulate the expression of jasmonate-responsive genes, such effects on the expression of JAZ genes were relatively weak (). Thus, MYC2 simultaneously induces the expression of both JAZ and JAM genes, with newly synthesized JAZ and JAM proteins negatively regulating jasmonate signaling. Although the three JAM transcription factors have redundant roles, JAM3 differs from JAM1 and JAM2 both in terms of its interactions with JAZ proteins and its expression profile after jasmonate treatment ().Citation21 Thus, the transcriptional activity of JAM3 to negatively regulate jasmonate-responsive genes might be regulated neither at the transcriptional level nor through interactions with JAZ proteins. In this study, we performed yeast two-hybrid screening and our results as well as the results of yeast two hybrid assay among JAM proteinsCitation24 indicated that JAM1 interacted with JAM3. One possibility is the formation of a heterodimer between JAM3 and JAM1 or JAM2 to modulate the transcriptional activity of JAM3. Given that JAM3 is constitutively expressed in almost all tissues (Genevestigator, https://www.genevestigator.com/gv/),Citation28 JAM3 protein may be stable in the presence or absence of stressors that cause the accumulation of JA-Ile. Given that the expression of JAM1 and JAM2 is quickly induced after MJ treatment,Citation21 it is plausible that newly synthesized JAM1 and JAM2 form heterodimers with JAM3 and that such interactions might reinforce the repression activity of the JAM protein complex.
Our results suggest that JAZ proteins are possible regulators of JAM proteins. Unlike MYC2, MYC3, and MYC4, JAM proteins are transcriptional repressors.Citation23,Citation24 Therefore, the effect of an interaction with JAZ proteins on the transcriptional repression activity of JAM proteins should be clarified. Further characterization of the protein interaction partners of JAM proteins is needed to understand the molecular mechanisms that regulate aspects of jasmonate signaling as controlled by JAM proteins.
Materials and Methods
Yeast two-hybrid screening
The JAM1 coding sequence was PCR-amplified with PrimeSTAR HS DNA Polymerase (TaKaRa, R010A) and Gateway-compatible primers by using a cDNA library from Arabidopsis thaliana seedlings (see Supplemental Table 1; primers 1 and 2). PCR products were cloned into pDONR/Zeo with a Gateway® BP Clonase® II enzyme mix (Invitrogen, 11789–020), and their sequences were verified. The JAM1 sequence was introduced into the destination low-copy yeast expression vector pDEST32 (with the GAL4 DNA binding domain [BD]) according to the manufacture’s instruction of Gateway® LR Clonase® enzyme mix (Invitrogen, 11791–019), and the sequence of the resulting plasmid was verified.
The pDEST32-JAM1 plasmid was transformed into yeast strain AH109. The prey cDNA library, Mate and Plate Library - Universal Arabidopsis (Clontech, 630487), was prepared in the plasmid pGADT7-Rec and transformed in yeast mating strain Y187. The transformed AH109 and the Y187 strain-based library were mixed with 1 mL and 5 mL of cultures, respectively (1 × 108 cells mL–1), and were incubated overnight in 45 mL of 2 × YPDA medium at 30 °C. Yeast diploids were selected by plating at 30 °C for 4 d on minimal medium SD lacking histidine (His), leucine (Leu), tryptophan (Trp), and adenine. Putative interacting partners of JAM1 that were isolated from this screen and their sequences were verified.
Yeast two-hybrid assay
cDNA clones that encode full-length bHLH transcription factors (JAM1 through JAM3 and MYC2), truncated derivatives of JAM1, and 12 full-length JAZ proteins were amplified with PrimeSTAR HS DNA Polymerase using Gateway-compatible primers (see Supplemental Table 1; For BP Gateway cloning). Those cDNA clones were introduced into the destination low-copy yeast expression vectors pDEST22 (with the Gal4 activation domain [AD]) and pDEST32. Truncated derivatives of JAM1 were used in combination with the destination low-copy yeast expression vector pDEST32. All constructs were checked by sequencing.
To assess protein interactions, the corresponding plasmids were cotransformed into Saccharomyces cerevisiae AH109 cells with a standard heat shock protocol.Citation25 Successfully transformed colonies were identified on yeast synthetic dropout medium that lacked Leu and Trp. At 3 d after transformation, yeast colonies were grown on yeast synthetic dropout medium lacking His, Leu, and Trp and supplemented with 5 mM 3-aminotriazole to test for protein interactions. Plates were incubated at 30 °C for 4 d. Cells were transformed with the empty Gateway vectors pDEST22 or pDEST32 to provide negative controls.
Alignment of the amino acid sequences of JAM1, JAM2, JAM3, MYC2, MYC3, and MYC4
The amino acid sequences of JAM1 and its closely related bHLH proteins were aligned using the ClustalW program (http://clustalw.ddbj.nig.ac.jp/).Citation29 Identical and similar amino acids are shaded black and gray, respectively, using the software GeneDoc ver 2.7 (http://www.nrbsc.org/gfx/genedoc/). Nuclear localization signals were predicted by cNLS Mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi).Citation30
Statistical analysis of GeneChip data
We used GeneChip data to obtain normalized intensities, the MJ/mock ratio for wild-type (WT) and jam×3 plants, and the jam×3/WT ratio for mock- and MJ-treated plants for all JAZ genes except JAZ11.Citation21 Then we performed a Tukey-Kramer multiple comparison test using the normalized intensities of JAZ genes and calculated the p values.
Quantitative reverse transcription PCR (qRT-PCR) analysis
Total RNA was isolated from 7-d-old Arabidopsis seedlings as described in Sasaki-Sekimoto et al.Citation21 cDNA was generated using ReverTra Ace® qRT-PCR RT Master Mix (TOYOBO, FSQ-201) and was used as a template for q RT-PCR analyses. We used THUNDERBIRD SYBR q RT-PCR Mix (TOYOBO, QPS-201) according to the manufacturer’s instructions. Gene-specific primers for ACTIN2 and JAZ1 are shown in Supplemental Table 1 (primers for qRT-PCR analysis).
Funding
This work was supported by Grants-in-Aid for Japan Society for the Promotion of Science Fellows (grant nos. 22–40129 and 19–8288 to Y.S.-S.) and The Ministry of Education, Culture, Sports, Science and Technology KAKENHI (grant no. 24228008 to K.S.). H.S. was supported by the Global Center of Excellence Program, from the Earth to “Earths,” at the Tokyo Institute of Technology and The University of Tokyo.
| Abbreviations: | ||
| bHLH | = | basic helix-loop-helix |
| JAM | = | JA-ASSOCIATED MYC2-LIKE |
| COI1 | = | CORONATINE INSENSITIVE1 |
| JA | = | Jasmonic acid |
| MJ | = | methyl jasmonate |
| qRT-PCR | = | quantitative reverse transcription PCR |
Additional material
Download Zip (50.8 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Akiko Ueno and Reiko Sakamoto for their assistance, as well as all the members of the Shirasu and Nakagami laboratories for their helpful discussions and technical support.
References
- Schilmiller AL, Howe GA. Systemic signaling in the wound response. Curr Opin Plant Biol 2005; 8:369 - 77; http://dx.doi.org/10.1016/j.pbi.2005.05.008; PMID: 15939667
- Browse J, Howe GA. New weapons and a rapid response against insect attack. Plant Physiol 2008; 146:832 - 8; http://dx.doi.org/10.1104/pp.107.115683; PMID: 18316637
- Browse J. Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol 2009; 60:183 - 205; http://dx.doi.org/10.1146/annurev.arplant.043008.092007; PMID: 19025383
- Sasaki Y, Asamizu E, Shibata D, Nakamura Y, Kaneko T, Awai K, Amagai M, Kuwata C, Tsugane T, Masuda T, et al. Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res 2001; 8:153 - 61; http://dx.doi.org/10.1093/dnares/8.4.153; PMID: 11572481
- Devoto A, Ellis C, Magusin A, Chang H-S, Chilcott C, Zhu T, Turner JG. Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol Biol 2005; 58:497 - 513; http://dx.doi.org/10.1007/s11103-005-7306-5; PMID: 16021335
- Sasaki-Sekimoto Y, Taki N, Obayashi T, Aono M, Matsumoto F, Sakurai N, Suzuki H, Hirai MY, Noji M, Saito K, et al. Coordinated activation of metabolic pathways for antioxidants and defence compounds by jasmonates and their roles in stress tolerance in Arabidopsis. Plant J 2005; 44:653 - 68; http://dx.doi.org/10.1111/j.1365-313X.2005.02560.x; PMID: 16262714
- Taki N, Sasaki-Sekimoto Y, Obayashi T, Kikuta A, Kobayashi K, Ainai T, Yagi K, Sakurai N, Suzuki H, Masuda T, et al. 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol 2005; 139:1268 - 83; http://dx.doi.org/10.1104/pp.105.067058; PMID: 16258017
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 2007; 19:2225 - 45; http://dx.doi.org/10.1105/tpc.106.048017; PMID: 17616737
- Pauwels L, Morreel K, De Witte E, Lammertyn F, Van Montagu M, Boerjan W, Inzé D, Goossens A. Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc Natl Acad Sci U S A 2008; 105:1380 - 5; http://dx.doi.org/10.1073/pnas.0711203105; PMID: 18216250
- Wasternack C. Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot 2007; 100:681 - 97; http://dx.doi.org/10.1093/aob/mcm079; PMID: 17513307
- Gfeller A, Dubugnon L, Liechti R, Farmer EE. Jasmonate biochemical pathway. Sci Signal 2010; 3:cm3; http://dx.doi.org/10.1126/scisignal.3109cm3; PMID: 20159849
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007; 448:666 - 71; http://dx.doi.org/10.1038/nature06006; PMID: 17637675
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007; 448:661 - 5; http://dx.doi.org/10.1038/nature05960; PMID: 17637677
- Staswick PE. JAZing up jasmonate signaling. Trends Plant Sci 2008; 13:66 - 71; http://dx.doi.org/10.1016/j.tplants.2007.11.011; PMID: 18261950
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004; 16:1938 - 50; http://dx.doi.org/10.1105/tpc.022319; PMID: 15208388
- Chico JM, Chini A, Fonseca S, Solano R. JAZ repressors set the rhythm in jasmonate signaling. Curr Opin Plant Biol 2008; 11:486 - 94; http://dx.doi.org/10.1016/j.pbi.2008.06.003; PMID: 18653378
- Katsir L, Chung HS, Koo AJK, Howe GA. Jasmonate signaling: a conserved mechanism of hormone sensing. Curr Opin Plant Biol 2008; 11:428 - 35; http://dx.doi.org/10.1016/j.pbi.2008.05.004; PMID: 18583180
- Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico J-M, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011; 23:701 - 15; http://dx.doi.org/10.1105/tpc.110.080788; PMID: 21335373
- Cheng Z, Sun L, Qi T, Zhang B, Peng W, Liu Y, Xie D. The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol Plant 2011; 4:279 - 88; http://dx.doi.org/10.1093/mp/ssq073; PMID: 21242320
- Niu Y, Figueroa P, Browse J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J Exp Bot 2011; 62:2143 - 54; http://dx.doi.org/10.1093/jxb/erq408; PMID: 21321051
- Sasaki-Sekimoto Y, Jikumaru Y, Obayashi T, Saito H, Masuda S, Kamiya Y, Ohta H, Shirasu K. Basic helix-loop-helix transcription factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3 are negative regulators of jasmonate responses in Arabidopsis. Plant Physiol 2013; 163:291 - 304; http://dx.doi.org/10.1104/pp.113.220129; PMID: 23852442
- Song S, Qi T, Fan M, Zhang X, Gao H, Huang H, Wu D, Guo H, Xie D. The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet 2013; 9:e1003653; http://dx.doi.org/10.1371/journal.pgen.1003653; PMID: 23935516
- Nakata M, Mitsuda N, Herde M, Koo AJ, Moreno JE, Suzuki K, Howe GA, Ohme-Takagi M. A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in arabidopsis. Plant Cell 2013; 25:1641 - 56; http://dx.doi.org/10.1105/tpc.113.111112; PMID: 23673982
- Nakata M, Ohme-Takagi M. Two bHLH-type transcription factors, JA-ASSOCIATED MYC2-LIKE2 and JAM3, are transcriptional repressors and affect male fertility. Plant Signal Behav 2013; 8:e26473; http://dx.doi.org/10.4161/psb.26473; PMID: 24056034
- Chini A, Fonseca S, Chico JM, Fernández-Calvo P, Solano R. The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J 2009; 59:77 - 87; http://dx.doi.org/10.1111/j.1365-313X.2009.03852.x; PMID: 19309455
- Melotto M, Mecey C, Niu Y, Chung HS, Katsir L, Yao J, Zeng W, Thines B, Staswick P, Browse J, et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J 2008; 55:979 - 88; http://dx.doi.org/10.1111/j.1365-313X.2008.03566.x; PMID: 18547396
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci U S A 2008; 105:7100 - 5; http://dx.doi.org/10.1073/pnas.0802332105; PMID: 18458331
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma 2008; 2008:420747.
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994; 22:4673 - 80; http://dx.doi.org/10.1093/nar/22.22.4673; PMID: 7984417
- Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A 2009; 106:10171 - 6; http://dx.doi.org/10.1073/pnas.0900604106; PMID: 19520826
