Abstract
Root system architecture (RSA) is developmentally controlled by genetic pathways and their interaction with various environmental cues, in particular soil conditions. One important player in shaping RSA is the hormone cytokinin, which acts as a negative regulator of root elongation and branching. The redundant roles of cytokinin metabolism and signaling genes of Arabidopsis thaliana in regulating early stages of lateral root formation has recently been shown and it has been proposed that this redundancy reflects a role in mediating different environmental cues. Here we report that the transcript levels of cytokinin genes in the root responds to changes in nutrient availability in distinct ways. IPT3, IPT5, CYP735A2, LOG5, and CKX4 are particularly responsive cytokinin metabolism genes, genes encoding different type-A response regulators and the transcriptions factor genes ARR10 and CRF6 are among the most responsive signaling genes. This finding supports the hypothesis that environmental cues operate through fine-tuned transcriptional regulation of cytokinin genes to modulate root development.
Plant roots respond to different nutrient deficiencies by changing their architecture in a way that will optimize nutrient uptake or help to withstand the stress.Citation1-Citation3 For example, phosphorous (P) deficiency in soils leads to an inhibition of primary root (PR) growth and an increase in number of first and second order of lateral roots (LR) aiming to explore the topsoil layers for phosphorous.Citation4,Citation5 In case of nitrogen (N) a deficiency stimulates the growth of a more exploratory root system with longer first order LR while higher N concentrations inhibit LR development by preventing meristem activation in LRPs.Citation5 This plasticity of roots has not only been shown to exist in responses to macronutrient alterations but also to some extent in those for micronutrients like iron (Fe), manganese (Mn), boran (B), and zinc (Zn). Apparently LR formation and development is most sensitive in responding to nutrient deficiencies.Citation5
LR initiation and development is tightly regulated by hormonal pathways. The hormone cytokinin (CK) is one important player acting as a negative regulator of LR formation and elongation. One of the hallmarks of the CK deficiency syndrome is an enlarged root system size which is mainly due to an increased number of lateral roots.Citation6-Citation8 Recently it has been shown that numerous CK metabolism and signaling genes act redundantly during early stages of LR development and it was hypothesized that this might reflect their function in mediating the consequences of changing environmental conditions.Citation9 Such a function would require that changing environmental cues would act on the activity of these genes or their products in order to alter the CK status for regulating RSA. Consistently, in a transcriptomic analysis a differential response of some of the cytokinin signaling genes in response to 4 different kinds of abiotic stress was found.Citation10 Here we show the analysis of the response of the complete set of CK metabolism and signaling genes to different environmental cues performed with transcriptome data which are publicly available in the Genevestigator database.Citation11 The following criteria were used for an arbitrary selection of microarray experiments available in the database and shown in and : 1) Preferentially the tested tissue were roots (whole seedlings or leaves were chosen in only a few cases when data from roots where not available or for comparison), 2) data recording the early consequences of changed environmental cues were preferred, and 3) only experiments using wild-type plants of accession Col-0 were considered unless specified otherwise.
Figure 1. Regulation of CK metabolism genes by different environmental cues. The changes in transcript abundance of the CK metabolism genes in response to different environmental cues or stresses were established with the Genevestigator databaseCitation11 (http://www.genevestigator.com/gv/index.jsp) and modified. The figure shows an arbitrary selection of experiments according to criteria outlined in the text. The gene expression responses were calculated as log2-ratios between the signal intensities from different stress or nutritional treatments compared with control or mock-treated samples. The resulting heatmap is color-coded as indicated and thus reflects the up- (red color) or downregulation (green color) of genes. The individual experiments are available from various repositories, such as Gene Expression OmnibusCitation25 or ArrayExpressCitation26 and can be retrieved by using their unique ID. Note that a probe for CKX7 is not present on the Affymetrix ATH1 chip. Abbreviations: d, days; vs., versus; Lz, longitudinal zone; Ws, Wassileskija; Col-4, Columbia-4.
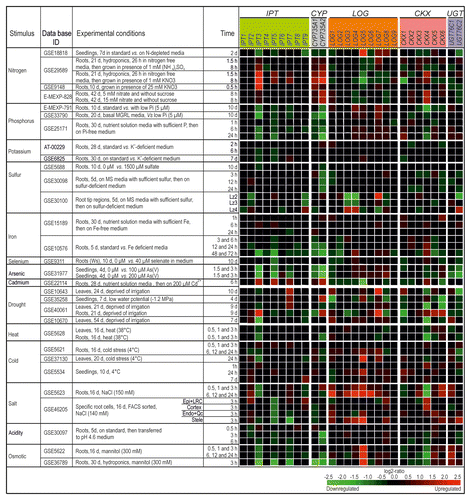
Figure 2. Regulation of CK signaling genes by different environmental cues. The changes in transcript abundance of CK signaling genes in response to different environmental cues or stresses was established with the Genevestigator databaseCitation11 (http://www.genevestigator.com/gv/index.jsp) and modified. The figure shows an arbitrary selection of experiments as is outlined in the text. The gene expression responses were calculated as log2-ratios between the signal intensities from different stress or nutritional treatments compared with control or mock-treated samples. The resulting heatmap is color coded as indicated and thus reflects the up- (red color) or downregulation (green color) of genes. The individual experiments are available from various repositories, such as Gene Expression OmnibusCitation25 or ArrayExpressCitation26 and can be retrieved by using their unique ID. Please note that only a single probe has been annotated on the Affymetrix ATH1 chip for both ARR13 and ARR21 and probes for CRF4 and CRF9 were not present. Abbreviations: d, days; vs., versus; Lz, longitudinal zone; Ws, Wassileskija; Col-4, Columbia-4.
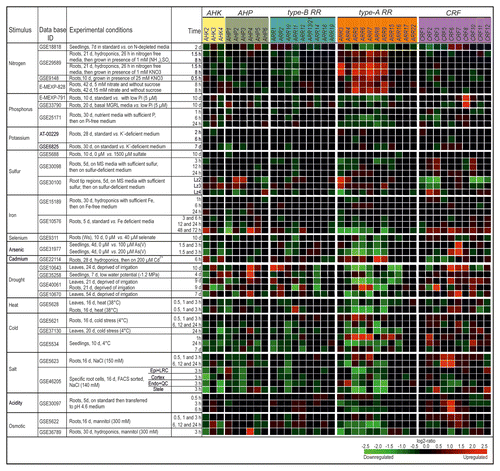
The analysis showed distinct changes in the transcript abundance of CK metabolism and signaling genes in response to differences in nutrient availability and different stresses. Generally, most of the conditions analyzed caused an altered transcript abundance of several CK genes. However, there was a wide range of responsiveness. For example, altered potassium concentrations caused almost no strong change of transcript abundance among the CK genes and also the early response to altered iron content was generally quite low. In contrast, addition of arsenic as well as drought and osmotic stress caused rapid changes of the transcript abundance of numerous CK metabolism and signaling genes. Other responses were more specific. For instance, altered phosphorus content in different experiments caused consistently a downregulation of the cytokinin-synthesizing IPT3 and CYP735A2 genes while the transcript levels of most other CK genes were not strongly altered ( and ).
Also a comparison of the responses to changes in the concentrations of 2 main forms of inorganic nitrogen, namely nitrate (NO3-) and ammonium (NH4+), revealed distinct responses of the CK genes. High nitrate upregulated the transcripts of CK metabolism genes IPT3Citation12,Citation13 and CYP735A2 while LOG5 was downregulated. In contrast, these genes did not respond to high ammonium in the medium (). Similarly, some CK signaling genes (type-A ARR genes ARR3, ARR5, and ARR7) were strongly responding to nitrate but not to ammonium ().
Recent abiotic stress experiments with CK deficient plants indicate that CKs are negative regulators of salt, drought, and osmotic stress signaling pathways and thereby control plant adaptation to various abiotic stresses.Citation14,Citation15 Arabidopsis roots exposed to salt treatments exhibit a general trend of repression of CK metabolism and signaling genes. Exceptions to this trend are some LOG (LOG1, LOG4, LOG5) and CRF (CRF5, CRF6) genes which were strongly upregulated in response to salt stress ( and ).
Also the analysis of the responsiveness of individual groups of CK genes reveals distinct reaction pattern. Among the CK synthesis genes IPT3 and IPT5 are responding most often to environmental cues. Interestingly, these 2 genes belong to those having a central role in regulating LR formation and growth.Citation9 Another particularly reactive gene is CYP735A2 which is one of the 2 CYP735A genes responsible for the production of trans-zeatin-type (tZ-type) CK which is transported from root to shoot.Citation16-Citation18 CYP735A2 is expressed specifically in the rootCitation18,Citation19 and mostly downregulated in response to stress. Interestingly, it has been shown recently that root-derived tZ-type cytokinins regulate shoot growth.Citation18 Thus, changed expression of CYP735A2 in response to stress might cause altered export of tZ-type CK from roots which in turn would regulate shoot growth in order to adapt to the stress condition. This idea is consistent with the decrease in tZ-type CK in xylem sap of drought-stressed plants as compared with well-watered control plants.Citation20,Citation21 The upregulation of CYP735A2 and IPT3 by high nitrate is also consistent with the known positive effect of nitrate on tZ-type CK levels in the xylem.Citation18,Citation22 Among the CK-degrading CKX genes the transcript level of the CKX4 gene is most often changed which has been reported previously to respond to different cues.Citation23
The transcripts encoding CK receptor genes respond relatively weak to environmental cues with the notable exception of salt stress which causes a reduction of AHK2 and CRE1/AHK4 transcripts in general and a specific reduction of AHK3 in root cortex cells ().Citation24 The spatially specific regulation of AHK3 was not detected in the analysis of whole roots indicating that probably also other cell type-specific responses to stress might have been diluted and thus remained unnoticed in transcriptomic studies of whole roots. Among the transcription factor genes operating downstream of the receptors ARR10 and CRF6 are the most responsive ones. Notably, ARR10 transcript levels are often downregulated by abiotic stress whereas CRF6 is upregulated () which indicates a bifurcated signaling pathway. Prominent response clusters are formed by the type-A ARR genes: Several members of this gene family are upregulated in roots in response to nitrogen and downregulated in response to arsenic, drought, heat, cold, salt, and osmotic stress. This indicates a lower CK status consistent with the above-described downregulation of the CK production as a stress response. Whether the altered transcript levels of genes encoding type-A ARR genes reflects only a lower CK status or is functionally relevant to locally modulate the response to the respective environmental cue deserves further investigation.
Collectively, the multiple and specific changes of CK gene transcript levels in response to different environmental cues support the idea that the hormone has a regulatory role in mediating information about altered environmental conditions leading to an adapted RSA according to the specific requirements.
| Abbreviations: | ||
| AHK | = | Arabidopsis histidine kinase |
| AHP | = | Arabidopsis histidine phosphotransmitter |
| ARR | = | Arabidopsis response regulator |
| CK | = | cytokinin |
| CRF | = | cytokinin response factor |
| CKX | = | cytokinin oxidase/dehydrogenase |
| IPT | = | isopentenyl transferase |
| LOG | = | Lonely guy |
| LR | = | lateral root |
| LRP | = | lateral root primordia |
| PR | = | primary root |
| RSA | = | root system architecture |
| tZ | = | trans-Zeatin |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We acknowledge funding from BMBF in the frame of the PLANT-KBBE program (ROOT- Root enhancement for crop improvement) and from DFG in the frame of SFB 973. The work of L.C. has been partly supported by a grant of the Dahlem Centre of Plant Sciences to T.S.
References
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L. The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 2003; 6:280 - 7; http://dx.doi.org/10.1016/S1369-5266(03)00035-9; PMID: 12753979
- Malamy JE. Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ 2005; 28:67 - 77; http://dx.doi.org/10.1111/j.1365-3040.2005.01306.x; PMID: 16021787
- Jung JKH, McCouch S. Getting to the roots of it: Genetic and hormonal control of root architecture. Front Plant Sci 2013; 4:186; http://dx.doi.org/10.3389/fpls.2013.00186; PMID: 23785372
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L. Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 2002; 129:244 - 56; http://dx.doi.org/10.1104/pp.010934; PMID: 12011355
- Gruber BD, Giehl RFH, Friedel S, von Wirén N. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol 2013; 163:161 - 79; http://dx.doi.org/10.1104/pp.113.218453; PMID: 23852440
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 2003; 15:2532 - 50; http://dx.doi.org/10.1105/tpc.014928; PMID: 14555694
- Werner T, Nehnevajova E, Köllmer I, Novák O, Strnad M, Krämer U, Schmülling T. Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 2010; 22:3905 - 20; http://dx.doi.org/10.1105/tpc.109.072694; PMID: 21148816
- Riefler M, Novak O, Strnad M, Schmülling T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 2006; 18:40 - 54; http://dx.doi.org/10.1105/tpc.105.037796; PMID: 16361392
- Chang L, Ramireddy E, Schmülling T. Lateral root formation and growth of Arabidopsis is redundantly regulated by cytokinin metabolism and signalling genes. J Exp Bot 2013; 64:5021 - 32; http://dx.doi.org/10.1093/jxb/ert291; PMID: 24023250
- Argueso CT, Ferreira FJ, Kieber JJ. Environmental perception avenues: the interaction of cytokinin and environmental response pathways. Plant Cell Environ 2009; 32:1147 - 60; http://dx.doi.org/10.1111/j.1365-3040.2009.01940.x; PMID: 19183294
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, et al. Genevestigator V3: A reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008; 420747.
- Takei K, Ueda N, Aoki K, Kuromori T, Hirayama T, Shinozaki K, Yamaya T, Sakakibara H. AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol 2004; a 45:1053 - 62; http://dx.doi.org/10.1093/pcp/pch119; PMID: 15356331
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 2004; 37:128 - 38; http://dx.doi.org/10.1046/j.1365-313X.2003.01945.x; PMID: 14675438
- Nishiyama R, Watanabe Y, Fujita Y, Le DT, Kojima M, Werner T, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Kakimoto T, et al. Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 2011; 23:2169 - 83; http://dx.doi.org/10.1105/tpc.111.087395; PMID: 21719693
- Jeon J, Kim NY, Kim S, Kang NY, Novák O, Ku SJ, Cho C, Lee DJ, Lee EJ, Strnad M, et al. A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J Biol Chem 2010; 285:23371 - 86; http://dx.doi.org/10.1074/jbc.M109.096644; PMID: 20463025
- Takei K, Yamaya T, Sakakibara H. Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin. J Biol Chem 2004; b 279:41866 - 72; http://dx.doi.org/10.1074/jbc.M406337200; PMID: 15280363
- Hirose N, Takei K, Kuroha T, Kamada-Nobusada T, Hayashi H, Sakakibara H. Regulation of cytokinin biosynthesis, compartmentalization and translocation. J Exp Bot 2008; 59:75 - 83; http://dx.doi.org/10.1093/jxb/erm157; PMID: 17872922
- Kiba T, Takei K, Kojima M, Sakakibara H. Side-chain modification of cytokinins controls shoot growth in Arabidopsis.. Dev Cell 2013; 27:452 - 61; http://dx.doi.org/10.1016/j.devcel.2013.10.004; PMID: 24286826
- Brenner WG, Schmülling T. Transcript profiling of cytokinin action in Arabidopsis roots and shoots discovers largely similar but also organ-specific responses. BMC Plant Biol 2012; 12:112; http://dx.doi.org/10.1186/1471-2229-12-112; PMID: 22824128
- Alvarez S, Marsh EL, Schroeder SG, Schachtman DP. Metabolomic and proteomic changes in the xylem sap of maize under drought. Plant Cell Environ 2008; 31:325 - 40; http://dx.doi.org/10.1111/j.1365-3040.2007.01770.x; PMID: 18088330
- Schachtman DP, Goodger JQD. Chemical root to shoot signaling under drought. Trends Plant Sci 2008; 13:281 - 7; http://dx.doi.org/10.1016/j.tplants.2008.04.003; PMID: 18467158
- Takei K, Sakakibara H, Taniguchi M, Sugiyama T. Nitrogen-dependent accumulation of cytokinins in root and the translocation to leaf: implication of cytokinin species that induces gene expression of maize response regulator. Plant Cell Physiol 2001; 42:85 - 93; http://dx.doi.org/10.1093/pcp/pce009; PMID: 11158447
- Werner T, Köllmer I, Bartrina I, Holst K, Schmülling T. New insights into the biology of cytokinin degradation. Plant Biol (Stuttg) 2006; 8:371 - 81; http://dx.doi.org/10.1055/s-2006-923928; PMID: 16807830
- Tran LS, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis.. Proc Natl Acad Sci U S A 2007; 104:20623 - 8; http://dx.doi.org/10.1073/pnas.0706547105; PMID: 18077346
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002; 30:207 - 10; http://dx.doi.org/10.1093/nar/30.1.207; PMID: 11752295
- Rocca-Serra P, Brazma A, Parkinson H, Sarkans U, Shojatalab M, Contrino S, Vilo J, Abeygunawardena N, Mukherjee G, Holloway E, et al. ArrayExpress: a public database of gene expression data at EBI. C R Biol 2003; 326:1075 - 8; http://dx.doi.org/10.1016/j.crvi.2003.09.026; PMID: 14744115
