Abstract
Xylan is the second most abundant polysaccharide in secondary walls of dicot plants and one of its structural features is the high degree of acetylation of xylosyl residues. In Arabidopsis, about 60% of xylosyl residues in xylan are acetylated and the biochemical mechanisms controlling xylan acetylation are largely unknown. A recent report by Yuan et al. (2013) revealed the essential role of a DUF231 domain-containing protein, ESKIMO1 (ESK1), in xylan acetylation in Arabidopsis as the esk1 mutation caused specific reductions in the degree of xylan 2-O or 3-O-monoacetylation and in the activity of xylan acetyltransferase. Interestingly, the esk1 mutation also resulted in an elevation of glucuronic acid (GlcA) substitutions in xylan. Since GlcA substitutions in xylan occur at the O-2 position of xylosyl residues, it is plausible that the increase in GlcA substitutions in the esk1 mutant is attributed to the reduction in acetylation at O-2 of xylosyl residues, which renders more O-2 positions available for GlcA substitutions. Here, we investigated the effect of removal of GlcA substitutions on the degree of xylan acetylation. We found that a complete loss of GlcA substitutions in the xylan of the gux1/2/3 triple mutant led to a significant increase in the degree of xylan acetylation, indicating that xylan acetyltransferases and glucuronyltransferases compete with each other for xylosyl residues for their acetylation or GlcA substitutions in planta. In addition, detailed structure analysis of xylan from the rwa1/2/3/4 quadruple mutant revealed that it had a uniform reduction of acetyl substitutions at different positions of the xylosyl residues, which is consistent with the proposed role of RWAs as acetyl coenzyme A transporters. The significance of these findings is discussed.
Plant biomass, which is mainly composed of cell walls, has been considered as a promising renewable source for production of second-generation liquid biofuels.Citation1 The major polysaccharides in cell walls of dicot plants that could be utilized for biofuel production are cellulose and xylan. Cellulose is made of a linear chain of glucose, which could be degraded by cellulolytic enzymes into glucose for fermentation into ethanol.Citation1 The structure of xylan is complex, having not only the backbone consisting of a linear chain of xylosyl residues but also substitutions of the xylosyl residues with glucuronic acid (GlcA), methylglucuronic acid (MeGlcA), arabinose, and acetyl groups depending on plant species.Citation2 Although xylose released from xylan could also be potentially used for fermentation into ethanol,Citation3 enzymatic degradation of xylan into xylose is difficult since the various substitutions in xylan negatively impact the digestion of xylan by xylanolytic enzymes.Citation4 Acetyl substitutions of xylan can occur at O-2 or O-3 of different xylosyl residues, at both O-2 and O-3 of the same xylosyl residue, and at O-3 of xylosyl residues substituted at O-2 with GlcA/MeGlcA.Citation5 The degree of acetylation of xylan has been analyzed in a number of plant species, including birch, beech, Populus, Eucalyptus, Paulownia, Acacia, Vitis, Arabidopsis, and maize, and found to vary from 0.35 to 0.61.Citation6-Citation11 Recent genetic and molecular analyses of 2 groups of Arabidopsis mutants, rwa and esk1, have provided the first glimpse into the biochemical mechanisms controlling xylan acetylation.Citation12-Citation14
Through analyzing genes that are activated by SND1, a secondary wall master transcriptional switch,Citation15,Citation16 Lee et al.Citation12 previously demonstrated that simultaneous mutations of 4 Arabidopsis RWA genes result in a reduction in xylan acetylation. Recently, Yuan et al.Citation14 have found that another SND1-activated gene, ESK1, is essential for the acetylation of xylan during secondary wall biosynthesis. The ESK1 gene is specifically expressed in secondary wall-forming cells, i.e., xylem and interfascicular fibers, and its encoded protein is localized in the Golgi. The esk1 mutation causes a reduction in secondary wall thickness and stem strength but has no effect on xylan content. Further chemical and structural analyses have revealed that the esk1 mutation results in a specific defect in 2-O- and 3-O-monoacetylation of xylosyl residues in xylan but not in 2,3-di-O-acetylation or 3-O-acetylation of xylosyl residues substituted at O-2 with GlcA. The defect in xylan acetylation in the esk1 mutant (also named tbl29) was also reported by Xiong et al.Citation13 Together with the finding that the esk1 mutation causes a reduction in xylan acetyltransferase activity, these results indicate that ESK1 is a strong candidate for xylan acetyltransferase that specifically mediates the 2-O- and 3-O-monoacetylation of xylosyl residues and also suggest that the 2,3-di-O-acetylation and 3-O-acetylation of xylosyl residues substituted at O-2 with GlcA are performed by other xylan acetyltransferase activities.
During the analysis of the distribution pattern of acetyl groups in esk1 xylan, it was noticed that the GlcA/MeGlcA substitutions in xylan were increased by 70% in esk1 compared with the wild type.Citation14 Since GlcA/MeGlcA is added at O-2 of xylosyl residues where acetylation also occurs, the increased GlcA/MeGlcA substitution in esk1 xylan is most likely attributed to the reduction in 2-O-acetylation rendering more O-2 positions available for addition of GlcA/MeGlcA groups. This finding suggests that the activities of xylan acetyltransferases and glucuronyltransferases may compete with each other for the O-2 positions of xylosyl residues in planta, i.e., higher acetyltransferase activities and lower glucuronyltransferase activities lead to more acetyl but less GlcA substitutions at O-2 of xylosyl residues and vice versa. To further substantiate this hypothesis, we investigated whether a reduction in GlcA substitutions in xylan, which leaves more O-2 positions available for substitutions, could increase the degree of acetyl substitutions. The xylan from the gux1/2/3 triple mutant is completely devoid of GlcA/MeGlcA substitutions,Citation17,Citation18 which provides an ideal material for this purpose. Acetylated xylans were isolated from the wild type and gux1/2/3 by DMSO extraction and analyzed for the patterns of acetyl substitutions by 1H nuclear magnetic resonance (NMR) spectroscopyCitation12 (). Acetylated xylans display resonances for the acetyl group around 2.15 ppm and those for all of the carbohydrate groups between 3.0 and 5.3 ppm.Citation5 The areas (integrals) of NMR signal peaks are directly proportional to the number of nuclei (e.g., protons) with the specific resonance frequencies (ppm). By integrating different NMR peaks, the ratio of protons with different resonance frequencies can be obtained to calculate the relative amounts of different structural groups.Citation19 Integrative analysis of the acetyl and total carbohydrate signal peaks of xylans revealed that the relative amount of the overall acetyl group in the gux1/2/3 xylan was increased by 23% compared with the wild-type xylan (). The NMR data was further confirmed by chemical analysisCitation12 showing that the gux1/2/3 xylan (86 mg of acetyl per g of total xylan) has about 19% more acetyl groups than the wild-type xylan (72 mg of acetyl per g of total xylan). As the observed increase in acetyl groups was measured using extracted xylans, it is unlikely that this increase could be a result of altered extraction properties of the gux1/2/3 cell wall polymers. Further detailed examination of the fingerprint regions of the 1H-NMR spectra showed that the wild-type xylan had characteristic resonances attributed to 2-O- and 3-O-monoacetylated xylosyl residues, 2,3-di-O-acetylated xylosyl residues, 3-O-monoacetylated xylosyl residues substituted at O-2 with GlcA/MeGlcA (). As expected, the gux1/2/3 xylan lacked the resonance signals corresponding to GlcA/MeGlcA and 3-O-monoacetylated xylosyl residues substituted at O-2 with GlcA/MeGlcA due to the absence of GlcA/MeGlcA substitutions. Integrative analysis showed that the resonance signals attributed to 2-O- and 3-O-monoacetylated xylosyl residues were significantly elevated in the gux1/2/3 xylan compared with the wild-type xylan (). These results demonstrate that a loss of xylan glucuronyltransferase activities as seen in the gux1/2/3 mutant, which results in a lack of GlcA substitutions in xylan, leads to an increase in the degree of xylan acetylation. This finding suggests that the degree of xylan acetylation or GlcA substitutions could be altered by a change in the relative activities of xylan acetyltransferases and glucuronyltransferases in planta. It further indicates that the relative activities of xylan acetyltransferases and glucuronyltransferases may be one of the factors that contribute to the variations in the degree of xylan acetylation seen in different plant species.Citation5-Citation11
Figure 1.1H nuclear magnetic resonance (NMR) spectra of acetylated xylans isolated from the wild type, gux1/2/3, and rwa1/2/3/4. Acetylated xylan was extracted with DMSOCitation6 and digested with β-endoxylanase M6 (Megazyme) to generate xylooligosaccharides, which were subsequently subject to structural analysis using NMR spectroscopy.Citation23 The resonance regions for acetyl groups and carbohydrate are marked.
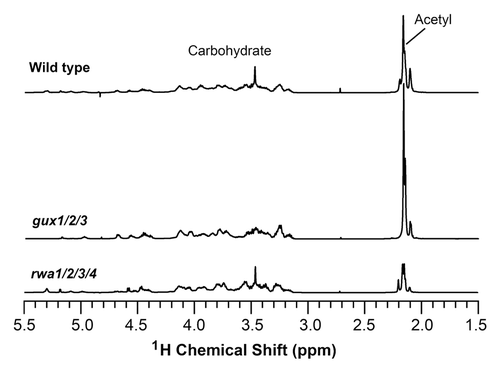
Table 1. Relative integrated values of acetyl groups and carbohydrates in DMSO-extracted xylans from Arabidopsis mutants and the wild type
Figure 2. Diagram of an acetylated xylooligomer from wild-type Arabidopsis xylan (A) and the fingerprint regions of the 1H NMR spectra of acetylated xylans of the wild type, gux1/2/3, and rwa1/2/3/4 (B). Resonances are labeled with the position of the assigned proton and the identity of structural fragments containing that proton. Resonances corresponding to non-acetylated xylosyl residues (Xyl), 2-O-acetylated xylosyl residues (Xyl-2Ac), 3-O-acetylated xylosyl residues (Xyl-3Ac), 2,3-di-O-acetylated xylosyl residues (Xyl-2,3Ac), 3-O-acetylated xylosyl residues substituted at O-2 with GlcA/MeGlcA (Xyl-3Ac-2GlcA), and GlcA/MeGlcA side chains are identified. The 1H NMR assignments were based on the NMR spectral data for the published xylan structure.Citation5,Citation14
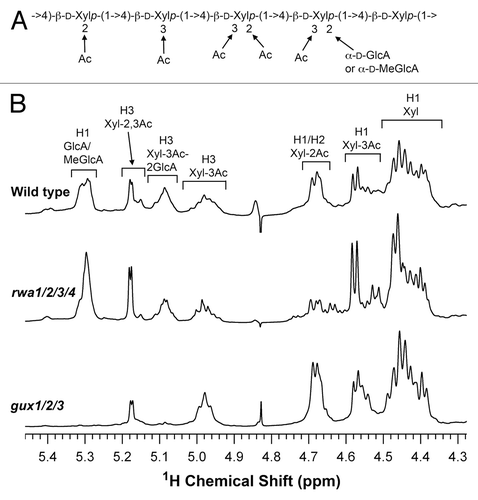
Table 2. Relative content of acetyl groups in xylans from Arabidopsis mutants and the wild type
We next examined whether the Arabidopsis rwa1/2/3/4 quadruple mutant, which is also defective in xylan acetylationCitation12 (; ), had any specific defects in the distribution pattern of acetyl groups on xylan. Integrative analysis of the resonances in the 1H-NMR spectra of acetylated xylan showed that the rwa1/2/3/4 mutant exhibited a reduction in all acetyl groups at different positions of the xylosyl residues (; ), a phenotype distinct from that caused by the esk1 mutation. This finding indicates that the rwa mutations affect a general process that is necessary for xylan acetylation at different positions of the xylosyl residues, which is consistent with the hypothesis that RWA proteins, which contain multiple transmembrane domains but lack a putative acetyltransferase domain,Citation20 are putative transporters of the acetyl donor, acetyl CoA.Citation21 A defect in the transport of acetyl CoA from the cytoplasm to the Golgi lumen would cause a reduction in the supply of acetyl CoA in the Golgi, which could subsequently lead to a uniform decrease in xylan acetylation at different positions of xylosyl residues. Whether RWAs indeed function as acetyl CoA transporters awaits further biochemical proof.
Our current study has demonstrated that a lack of GlcA substitutions in xylan as seen in the gux1/2/3 mutant can lead to an increase in the degree of xylan acetylation. Since the acetyl group is released in the form of acetate during during pretreatments of biomass for biofuel production, which has been shown to be inhibitory to microorganisms used for sugar fermentation,Citation22 the elevated degree of xylan acetylation in the gux1/2/3 mutant may partially offset the potential benefit of improved alkaline extractability of the mutant xylan.Citation17 As exemplified in this study, a change in one kind of modification in cell wall polysaccharides may lead to other alterations, and therefore, it is pivotal to have a complete understanding of the biochemical mechanisms controlling cell wall biosynthesis in order to custom-design plant biomass better suited for biofuel production. In particular, further understanding of the biochemical mechanisms controlling xylan acetylation will undoubtedly provide knowledge basis for genetic manipulation of plant biomass with reduced degrees of cell wall acetylation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was funded by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy through Grant DE-FG02–03ER15415.
References
- Carroll A, Somerville C. Cellulosic biofuels. Annu Rev Plant Biol 2009; 60:165 - 82; http://dx.doi.org/10.1146/annurev.arplant.043008.092125; PMID: 19014348
- Timell TE. Recent progress in the chemistry of wood hemicelluloses. Wood Sci Technol 1967; 1:45 - 70; http://dx.doi.org/10.1007/BF00592255
- Lee H, Biely P, Latta RK, Barbosa MF, Schneider H. Utilization of xylan by yeasts and its conversion to ethanol by Pichia stipitis strains. Appl Environ Microbiol 1986; 52:320 - 4; PMID: 16347130
- Selig MJ, Adney WS, Himmel ME, Decker SR. The impact of cell wall acetylation on corn stover hydrolysis by cellulolytic and xylanolytic enzymes. Cellulose 2009; 16:711 - 22; http://dx.doi.org/10.1007/s10570-009-9322-0
- Teleman A, Lundqvist J, Tjerneld F, Stålbrand H, Dahlman O. Characterization of acetylated 4-O-methylglucuronoxylan isolated from aspen employing 1H and 13C NMR spectroscopy. Carbohydr Res 2000; 329:807 - 15; http://dx.doi.org/10.1016/S0008-6215(00)00249-4; PMID: 11125823
- Teleman A, Tenkanen M, Jacobs A, Dahlman O. Characterization of O-acetyl-(4-O-methylglucurono)xylan isolated from birch and beech. Carbohydr Res 2002; 337:373 - 7; http://dx.doi.org/10.1016/S0008-6215(01)00327-5; PMID: 11841818
- Evtuguin DV, Tomás JL, Silva AM, Neto CP. Characterization of an acetylated heteroxylan from Eucalyptus globulus Labill. Carbohydr Res 2003; 338:597 - 604; http://dx.doi.org/10.1016/S0008-6215(02)00529-3; PMID: 12644372
- Gonçalves VM, Evtuguin DV, Domingues MR. Structural characterization of the acetylated heteroxylan from the natural hybrid Paulownia elongata/Paulownia fortunei.. Carbohydr Res 2008; 343:256 - 66; http://dx.doi.org/10.1016/j.carres.2007.11.002; PMID: 18039538
- Naran R, Black S, Decker SR, Azadi P. Extraction and characterization of native heteroxylans from delignified corn stover and aspen. Cellulose 2009; 16:661 - 75; http://dx.doi.org/10.1007/s10570-009-9324-y
- Marques G, Gutierrez A, del Rio JC, Evtuguin DV. Acetylated heteroxylan from Agave sisalana and its behavior in alkaline pulping and TCF/ECF bleaching. Carbohydr Polym 2010; 81:517 - 23; http://dx.doi.org/10.1016/j.carbpol.2010.02.043
- Prozil SO, Costa EV, Evtuguin DV, Lopes LP, Domingues MR. Structural characterization of polysaccharides isolated from grape stalks of Vitis vinifera L. Carbohydr Res 2012; 356:252 - 9; http://dx.doi.org/10.1016/j.carres.2012.02.001; PMID: 22398255
- Lee C, Teng Q, Zhong R, Ye Z-H. The four Arabidopsis reduced wall acetylation genes are expressed in secondary wall-containing cells and required for the acetylation of xylan. Plant Cell Physiol 2011; 52:1289 - 301; http://dx.doi.org/10.1093/pcp/pcr075; PMID: 21673009
- Xiong G, Cheng K, Pauly M. Xylan O-acetylation impacts xylem development and enzymatic recalcitrance as indicated by the Arabidopsis mutant tbl29.. Mol Plant 2013; 6:1373 - 5; http://dx.doi.org/10.1093/mp/sst014; PMID: 23340742
- Yuan Y, Teng Q, Zhong R, Ye Z-H. The Arabidopsis DUF231 domain-containing protein ESK1 mediates 2-O- and 3-O-acetylation of xylosyl residues in xylan. Plant Cell Physiol 2013; 54:1186 - 99; http://dx.doi.org/10.1093/pcp/pct070; PMID: 23659919
- Zhong R, Lee C, Ye Z-H. Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol 2010; 152:1044 - 55; http://dx.doi.org/10.1104/pp.109.148270; PMID: 19965968
- Zhong R, McCarthy RL, Lee C, Ye Z-H. Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol 2011; 157:1452 - 68; http://dx.doi.org/10.1104/pp.111.181354; PMID: 21908685
- Mortimer JC, Miles GP, Brown DM, Zhang Z, Segura MP, Weimar T, Yu X, Seffen KA, Stephens E, Turner SR, et al. Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. Proc Natl Acad Sci U S A 2010; 107:17409 - 14; http://dx.doi.org/10.1073/pnas.1005456107; PMID: 20852069
- Lee C, Teng Q, Zhong R, Ye Z-H. Arabidopsis GUX proteins are glucuronyltransferases responsible for the addition of glucuronic acid side chains onto xylan. Plant Cell Physiol 2012; 53:1204 - 16; http://dx.doi.org/10.1093/pcp/pcs064; PMID: 22537759
- Teng Q. Structural Biology: Practical NMR Applications. 2nd ed. New York: Springer, 2013
- Janbon G, Himmelreich U, Moyrand F, Improvisi L, Dromer F. Cas1p is a membrane protein necessary for the O-acetylation of the Cryptococcus neoformans capsular polysaccharide. Mol Microbiol 2001; 42:453 - 67; http://dx.doi.org/10.1046/j.1365-2958.2001.02651.x; PMID: 11703667
- Gille S, de Souza A, Xiong G, Benz M, Cheng K, Schultink A, Reca IB, Pauly M. O-acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. Plant Cell 2011; 23:4041 - 53; http://dx.doi.org/10.1105/tpc.111.091728; PMID: 22086088
- Helle S, Cameron D, Lam J, White B, Duff S. Effect of inhibitory compounds found in biomass hydrolysates on growth and xylose fermentation by a genetically engineered strain of S. cerevisiae.. Enzyme Microb Technol 2003; 33:786 - 92; http://dx.doi.org/10.1016/S0141-0229(03)00214-X
- Teng Q, Ekman DR, Huang W, Collette TW. Push-through direct injection NMR: an optimized automation method applied to metabolomics. Analyst 2012; 137:2226 - 32; http://dx.doi.org/10.1039/c2an16251b; PMID: 22434060
