Abstract
Xylan is the major hemicellulose present in both primary and secondary cell walls of rice vegetative tissues. Since xylan is one of the factors contributing to biomass recalcitrance, understanding how xylan is synthesized in rice will potentially provide tools to modify grass biomass composition better suited for biofuel production. Studies of xylan biosynthesis in Arabidopsis have revealed that family GT43 glycosyltransferases, which form 2 functionally nonredundant groups, IRX9/IRX9 homolog and IRX14/IRX14 homolog, are required for xylan backbone elongation. The rice genome harbors 10 genes encoding family GT43 members and it is currently unknown whether they are all involved in xylan biosynthesis. In this report, we performed biochemical analysis of xylan xylosyltransferase activity in rice stem microsomes and investigated the roles of 4 representative rice GT43 members, OsGT43A (LOC_Os05 g03174), OsGT43E (LOC_Os05 g48600), OsGT43H (LOC_Os04 g01280), and OsGT43J (LOC_Os06 g47340), in xylan biosynthesis. OsGT43 proteins were shown to be localized in the Golgi, where xylan biosynthesis occurs. Complementation analysis by expression of OsGT43s in Arabidopsis irx9 and irx14 mutants demonstrated that OsGT43A and OsGT43E but not OsGT43H and OsGT43J were able to rescue the mutant phenotypes conferred by the irx9 mutation, including defective stem mechanical strength, vessel morphology, xylan content, GlcA side chains, xylan chain length, and xylosyltransferase activity. On the other hand, OsGT43J but not OsGT43A, OsGT43E, and OsGT43H restored the defective xylan phenotype in the irx14 mutant. These results indicate that the rice GT43 family evolved to retain the involvement of 2 functionally nonredundant groups, OsGT43A and OsGT43E (IRX9 homologs) vs. OsGT43J (an IRX14 homolog), in xylan backbone biosynthesis.
Keywords: :
Introduction
Cellulosic biomass from grasses, such as rice (Oryza sativa), corn (Zea mays), switchgrass (Panicum virgatum), and Miscanthus (Miscanthus giganteus), has been considered to be a promising source of renewable bioenergy for bioethanol production.Citation1 However, the cell wall, which is the primary constituent of cellulosic biomass, is intrinsically recalcitrant to enzymatic degradation, thus hampering the efficient utilization of cellulosic biomass for biofuel production. Lignin and xylan are the 2 major polymers in grass cell walls contributing to biomass recalcitrance, i.e., they form physical barriers to block cellulolytic enzymes from hydrolyzing cellulose into fermentable sugars.Citation2 To overcome biomass recalcitrance, it is crucial to uncover the molecular and biochemical mechanisms controlling the biosynthesis and assembly of cell wall components. The knowledge thus gained could be applied to modify cell wall composition tailored for biofuel production.
Xylan is the major crosslinking (noncovalently and covalently) hemicellulose in both primary and secondary cell walls of grasses.Citation2 It is made of a linear chain of β-1,4-linked xylosyl residues, some of which are substituted by α-1,2/α-1,3-linked arabinose, α-1,2-linked glucuronic acid (GlcA), α-1,2-linked 4-O-methylglucuronic acid (MeGlcA), and acetylated at C-2 or C-3.Citation3 Different from the xylan in dicots, grass xylan has several unique features. In addition to arabinose as its predominant side chains, grass xylan covalently crosslinks with lignin via ferulic acid residues and has xylosyl substitution at O-2 of the arabinosyl side chains.Citation4 Bioinformatic studies and transcriptome profiling analyses have revealed that a number of genes encoding families GT43, GT47, and GT61 glycosyltransferases and acyl-CoA transferases are implicated in xylan biosynthesis in grasses.Citation5-Citation7 Among them, one GT61 member from wheat and its rice close homologs were proven to mediate the addition of arabinosyl residues onto xylanCitation8 and another GT61 member from rice is involved in the xylosyl substitution at O-2 of the arabinosyl side chains in xylan.Citation9 Four acyl-CoA transferases from rice were proposed to be putative arabinoxylan feruloyltransferases since downregulation of their expression in transgenic rice resulted in a moderate reduction (19%) of ferulate in cell walls.Citation10 Intriguingly, overexpression of another acyl-CoA transferase that is a close homolog of these putative arabinoxylan feruloyltransferases in transgenic rice caused an increase in p-coumarate but not ferulate in cell walls, which led the authors to suggest that the protein is an arabinoxylan coumaroyltransferase.Citation11 Genes responsible for the addition and methylation of the GlcA side chains in xylan and those involved in xylan acetylation have not yet been characterized in grasses. In Arabidopsis, it has been demonstrated that 3 GT8 genes encode glucuronyltransferases required for GlcA substitution in xylan,Citation12-Citation14 3 DUF579 domain-containing proteins are glucuronoxylan methyltransferases catalyzing the 4-O-methylation of the GlcA side chains,Citation15,Citation16 and 4 RWA genes and one DUF231 domain-containing protein, ESK1/TBL29, are implicated in xylan acetylation.Citation17-Citation19
The biochemical mechanism controlling xylan backbone elongation appears to be much more complicated than expected. In contrast to the biosynthesis of secondary wall cellulose in which 3 functionally nonredundant family GT2 cellulose synthases are involved,Citation20 xylan backbone biosynthesis entails glycosyltransferases from both GT43 and GT47 families.Citation21-Citation25 It has been demonstrated that the 4 Arabidopsis GT43 members form 2 functionally nonredundant groups, IRX9/IRX9 homolog and IRX14/IRX14 homolog, both of which are required for xylan backbone biosynthesis.Citation26,Citation27 Biochemical analysis has further revealed that IRX9 and IRX14 are xylosyltransferases that act cooperatively in the elongation of the xylan backbone.Citation28 Simultaneous mutations of 2 Arabidopsis GT47 genes, IRX10 and IRX10L, also result in defects in xylan backbone elongation, indicating their essential role in xylan backbone biosynthesis.Citation24,Citation25 It is currently unknown why the elongation of β-1,4-linked xylosyl residues requires glycosyltransferases from 2 different GT families.
Studies of xylan biosynthesis in grasses have also revealed the involvement of both GT43 and GT47 glycosyltransferases in xylan biosynthesis.Citation29,Citation30 However, analysis of the protein complex possessing xylan xylosyltransferase activity in wheat only identified one GT43 member that is a close homolog of Arabidopsis IRX14.Citation29 Thus, it is unclear whether the xylan biosynthesis in grasses requires 2 functionally nonredundant GT43 members as in Arabidopsis. Since an expansion of the GT43 family occurred in grass species such as rice, it necessitates investigation on whether there is functional diversification in the GT43 family in grasses. In this report, we studied the biochemical properties of xylan xylosyltransferase activity in rice stem microsomes and investigated the functions of rice GT43 members in xylan biosynthesis. Our results revealed both conservation and divergence in the functional roles of rice family GT43 members in xylan biosynthesis.
Results
Structure of rice xylan and biochemical properties of rice xylan xylosyltransferase activity
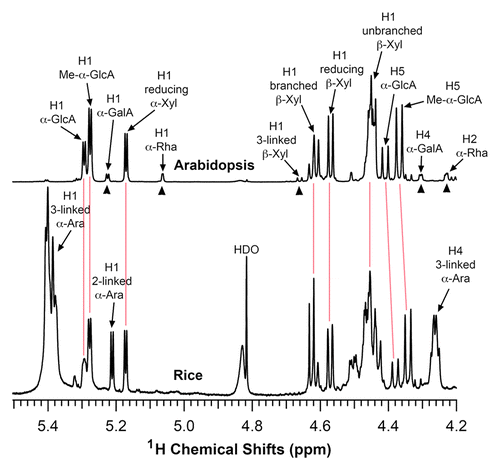
To investigate the biochemical mechanism controlling xylan biosynthesis in rice, we first analyzed the structure of xylan isolated from rice stems. KOH-extracted xylan was first digested with β-1,4-endoxylanase and its structure was subsequently determined using nuclear magnetic resonance (NMR) spectroscopy (). Rice xylan exhibited the same resonances attributed to the backbone xylosyl residues (H1 of branched and unbranched at 4.62 and 4.46 ppm, respectively) and the GlcA side chains (H1 of GlcA and MeGlcA at 5.29 and 5.27 ppm, respectively) as Arabidopsis xylan. However, the resonances characteristic of the Arabidopsis xylan reducing end tetrasaccharide sequence, β-d-Xyl-(1→3)-α-l-Rha-(1→2)-α-d-GalA-(1→4)-d-Xyl (H1 of 3-linked β-D-Xyl, H1 of α-L-Rha, H1 of α-D-GalA, H2 of α-L-Rha, and H4 of α-D-GalA), was not evident in rice xylan, indicating that either rice xylan does not possess the reducing end tetrasaccharide sequence or the amount is too low to be detected. The most prominent difference between rice and Arabidopsis xylans is that the resonances characteristic of the arabinose side chains (H1 3-linked at 5.4 ppm, H4 3-linked at 4.26 ppm, and H1 2-linked at 5.21 ppm) were only present in rice xylan. The structural analysis demonstrated that similar to xylan in other grass species,Citation31,Citation32 xylan from rice stems consists of a linear chain of β-1,4-linked xylosyl residues, some of which are substituted with side chains of α-1,2-linked GlcA, α-1,2-linked MeGlcA, and α-1,2/1,3-linked arabinose. The 1H-NMR spectrum shows clearly the resonances of different structural groups of rice stem xylan, which could be used as a reference for future study of xylan in transgenic rice plants with altered xylan structure.
Figure 1. Structural analysis of rice xylan by NMR spectroscopy. Xylooligosaccharides generated by β-endoxylanase digestion of alkaline-extracted xylan were subjected to 1H-NMR analysis. Resonances are labeled with the position of the assigned proton and the identity of the residue containing that proton. The NMR spectrum of Arabidopsis xylan was included for comparison. The proton resonances shared by both Arabidopsis and rice xylans are marked by vertical red lines. The resonances of H1 of α-D-GalA, H1 of α-L-Rha, H1 of 3-linked β-D-Xyl, H4 of α-D-GalA, and H2 of α-L-Rha (arrow heads) from the reducing end tetrasaccharide sequence of Arabidopsis xylan was not observed in rice xylan. Note the predominant resonances corresponding to 2- and 3-linked α-arabinose in rice xylan, which is absent in Arabidopsis xylan. HDO, hydrogen deuterium oxide.
We next examined the xylan xylosyltransferase activity in microsomes isolated from rice stems. It was found that rice microsomes possessed a xylosyltransferase activity capable of catalyzing the transfer of xylosyl residues from 14C-labeled UDP-xylose onto the exogenous xylotetraose (Xyl4) acceptor in a concentration-dependent manner (). No xylosyltransferase activity was detected in the absence of the exogenous Xyl4 acceptor, which is different from the xylosyltransferase activity of wheat microsomes in which no exogenous acceptors are required (Zeng et al., 2010). Under the conditions used, the xylosyltransferase activity of rice microsomes was time-dependent reaching its maximum after 90 min incubation () and was also dependent on microsomal protein concentration ().
Figure 2. Biochemical properties of xylan xylosyltransferase activity in microsomes from rice stems. The xylosyltransferase activity was assayed by incubating microsomes with the UDP-[14C]-xylose donor and the Xyl4 acceptor for 30 min unless otherwise indicated. The activity (CPM) was measured by the transfer of the radiolabeled xylosyl group onto the acceptor. All assays were repeated twice and the data are means ± SE of assays from 3 independently isolated microsomes. (A) Dependence of the xylosyltransferase activity on the Xyl4 acceptor concentration. (B) Time course of the transfer of the radiolabeled xylosyl residues onto the acceptor by the xylosyltransferase activity. (C) The xylosyltransferase activity is protein concentration-dependent.
![Figure 2. Biochemical properties of xylan xylosyltransferase activity in microsomes from rice stems. The xylosyltransferase activity was assayed by incubating microsomes with the UDP-[14C]-xylose donor and the Xyl4 acceptor for 30 min unless otherwise indicated. The activity (CPM) was measured by the transfer of the radiolabeled xylosyl group onto the acceptor. All assays were repeated twice and the data are means ± SE of assays from 3 independently isolated microsomes. (A) Dependence of the xylosyltransferase activity on the Xyl4 acceptor concentration. (B) Time course of the transfer of the radiolabeled xylosyl residues onto the acceptor by the xylosyltransferase activity. (C) The xylosyltransferase activity is protein concentration-dependent.](/cms/asset/212834c1-51e7-49d0-9d17-17d94fe1dbc6/kpsb_a_10927809_f0002.gif)
The xylan xylosyltransferase activity exhibited by the rice stem microsomes were further assessed by HPLC using the fluorescent anthranilic acid (AA)-labeled xyolooligomers of different lengths as acceptors (). The acceptors, xylooligomers, used for the assay were purchased from Megazyme and they are free from contamination as verified by HPLC. No xylosyl transfer was detected when the monomer AA-Xyl was used as the acceptor and a low level of xylosyl transfer was observed when AA- Xyl2 was used. Up to 4 xylosyl residues were able to be efficiently transferred onto the acceptors ranging from Xyl3 to Xyl6 by the xylosyltransferase activity of rice microsomes. No addition of xylosyl residues onto the xylooligomer acceptors was observed when UDP-xylose was absent (). As the elongation of xylooligomers depends on the presence of the xylosyl donor, UDP-xylose, and the reducing end of the xylooligomers is blocked by anthranilic acid, it is unlikely that this observed addition of xylosyl residues onto the xylooligomer acceptors could be mediated by putative xylan transglycosylase activities. It should be pointed out that although the rice microsomes exhibit a xylosyltransferase activity that is able to successively transfer up to 4 xylosyl residues onto the xylooligomer acceptors (), prolonged incubation of the reactions did not generate reaction products with a longer chain length. One possible explanation is that long xylooligosaccharides without GlcA side chains and acetyl modifications are insoluble as shown in the alkaline-extracted xylan from the gux1 gux 2 double mutant,Citation12 and thus they could not be produced. The other possible explanation is that a xylosidase activity present in rice microsomes degrades reaction products (see below), which prevents the accumulation of xylooligosaccharides with a longer chain length. The data shown here are in agreement with an early report showing that wheat microsomes possess a xylosyltransferase activity that adds up to 2 xylosyl residues onto the xylooligomer acceptor.Citation33
Figure 3. Xylooligomers of various lengths as acceptors for the xylosyltransferase activity of rice microsomes. The xylosyltransferase activity was assayed by incubating rice stem microsomes with UDP-xylose and the acceptor Xyl1-AA, Xyl2-AA, Xyl3-AA, Xyl4-AA, Xyl5-AA, or Xyl6-AA, and the reaction products were analyzed by reverse-phase HPLC and detected for fluorescent signals. The number at each peak denotes the number of xylosyl residues for the corresponding xylooligomer. A chromatogram of standard Xyl1–6-AA is shown at the top of each column for comparison of the xylooligomer peaks.
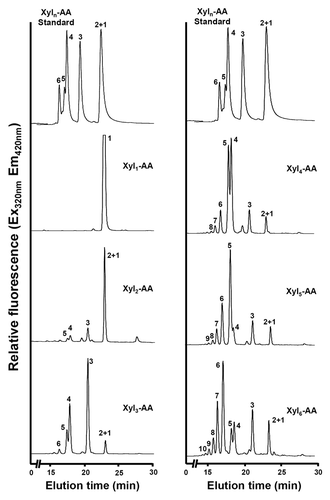
Figure 4. Digestion of the xylosyltransferase-catalyzed reaction products by β-1,4-xylosidase. Microsomes were incubated with the fluorescent Xyl6-AA acceptor and UDP-xylose. The reaction products were digested with β-1,4-xylosidase and then analyzed by reverse-phase HPLC. (A) The control reaction without UDP-xylose showing the Xyl6-AA acceptor peak. (B) Rice microsomes possess the xylosyltransferase activity capable of adding xylosyl residues onto the Xyl6-AA acceptor peak. (C) β-1,4-Xylosidase digestion of the reaction products from (B) results in their degradation into Xyl1-AA and Xyl2-AA, indicating that the reaction products are β-1,4-linked. The number at each peak denotes the number of xylosyl residues for the corresponding xylooligomer.
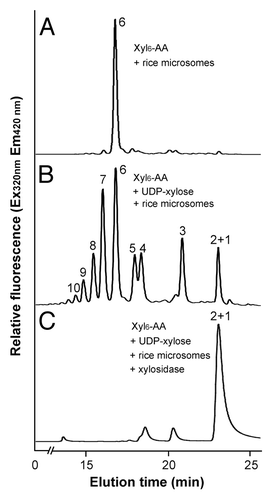
It was of note that the reaction products not only consisted of xylooligomers longer than the acceptor but also contained shorter xylooligomers (), indicating that in addition to the xylosyltransferase activity catalyzing the addition of xylosyl residues, the rice microsomes contain a xylosidase-like activity that removes xylosyl residues from the acceptors. It remains to be investigated what physiological roles this xylosidase-like activity may play during xylan biosynthesis in rice. Little xylosidase-like activity was observed in xylan xylosyltransferase activity assays with microsomes from Arabidopsis and poplar.Citation22,Citation34
The reaction products, xylooligosaccharides, generated by rice xylosyltransferase activity were further verified for their sugar linkage by digestion with β-(1,4)-xylosidase (). HPLC analysis of digested products showed that treatment of the microsome-catalyzed reaction products () with β-(1,4)-xylosidase led to their hydrolysis into xylose or xylobiose (), demonstrating that the reaction products are made of β-1,4-linked xylosyl residues.
Family GT43 glycosyltransferases in rice
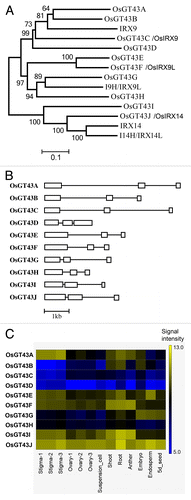
In Arabidopsis, all 4 members of the GT43 family are xylosyltransferases involved in xylan backbone elongation.Citation21-Citation23,Citation26-Citation28 There exist 10 GT43 genes in the rice genome and phylogenetic analysis showed that there is a significant expansion of OsGT43 members that are closely grouped together with Arabidopsis IRX9 and I9H/IRX9L (). All the rice GT43 genes have a similar exon-intron organization, i.e., they contain 3 exons and 2 introns (). The expression of all these rice GT43 genes, except OsGT43C and OsGT43D, was evident in shoots and roots (). We selected 4 representative members, OsGT43A, OsGT43E, OsGT43H, and OsGT43J, which are positioned at different nodes in the GT43 phylogenetic tree, for further functional analysis. Among them, OsGT43A and OsGT43E are phylogenetically closer to IRX9, OsGT43H closer to I9H/IRX9L, and OsGT43J closer to IRX14.
Figure 5. Phylogenetic analysis of rice and Arabidopsis family GT43 proteins (A), exon-intron organization (B) and expression patterns (C) of OsGT43 genes. (A) The GT43 amino acid sequences were aligned using ClustalW and their phylogenetic relationship was analyzed using the neighbor-joining method in MEGA5.2 (Tamura et al., 2011). Bootstrap values resulted from 1,000 replicates are shown at the nodes. (B) Open boxes and lines denote exons and introns, respectively. The bar scale shows the number of nucleotides. (C) The expression of OsGT43 genes in various tissues shown as the heat map was from Li et al.Citation50 Shoot and root tissues are from 2-week-old shoots and roots of rice (Oryza sativa Japonica Nipponbare).
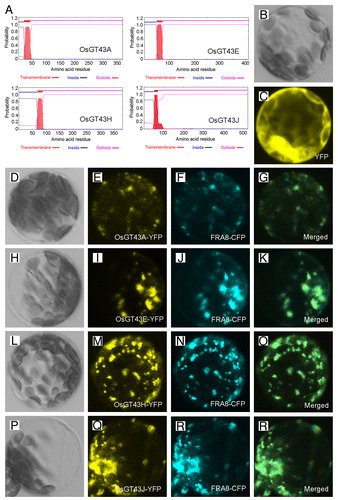
To investigate whether rice GT43 proteins are involved in xylan biosynthesis, we first examined their subcellular localization. The OsGT43A/E/H/J proteins were predicted to be type II membrane proteins with one transmembrane helix at the N-terminus () and when expressed in Arabidopsis protoplasts, the yellow fluorescence protein (YFP)-tagged OsGT43 proteins were co-localized with the cyan fluorescence protein (CFP)-tagged FRAGILE FIBER8 (FRA8) (), a GT47 glycosyltransferase known to be localized in the Golgi.Citation35 The control protoplasts expressing YFP alone exhibited signals throughout the cytoplasm (). These results demonstrates that the OsGT43 proteins are localized in Golgi, where xylan biosynthesis occurs.
Figure 6. Subcellular localization of OsGT43 proteins. Fluorescent protein-tagged fusion proteins were expressed in Arabidopsis protoplasts, and the signals were visualized with a laser confocal microscope. (A) OsGT43A, OsGT43E, OsGT43H and OsGT43J are membrane proteins as predicted by the TMHMM2.0 program. Inside, the cytoplasmic side of the membrane; outside, the noncytoplasmic side of the membrane. (B) and (C) An Arabidopsis leaf protoplast (B) expressing YFP alone showing the fluorescent signals throughout the cytoplasm (C). (D) to (G) An Arabidopsis protoplast (D) co-expressing OsGT43A-YFP (E) and the Golgi-localized FRA8-CFP (F). (H) to (K) An Arabidopsis protoplast (H) co-expressing OsGT43E-YFP (I) and FRA8-CFP (J). (L) to (O) An Arabidopsis protoplast (L) co-expressing OsGT43H-YFP (M) and FRA8-CFP (N). (P) to (S) An Arabidopsis protoplast (P) co-expressing OsGT43J-YFP (Q) and FRA8-CFP (R). Note the superimposition of the fluorescent signals of OsGT43-YFP with FRA8-CFP (G, K, O, and S).
OsGT43A and OsGT43E but not OsGT43H and OsGT43J are functional orthologs of Arabidopsis IRX9
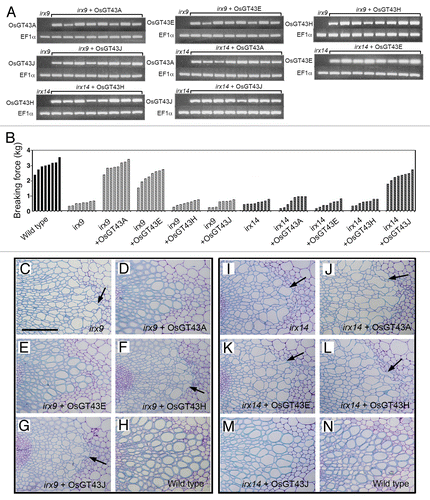
Arabidopsis GT43 members form 2 functionally non-redundant groups, IRX9/IRX9 homolog and IRX14/IRX14 homolog, both of which are required for xylan biosynthesis.Citation26,Citation27 To investigate the functional roles of OsGT43 members in xylan biosynthesis, we examined the ability of the 4 representative OsGT43 genes, OsGT43A, OsGT43E, OsGT43H, and OsGT43J, to complement the xylan defects conferred by the irx9 and irx14 mutations in Arabidopsis. The full-length OsGT43 cDNAs driven by the cauliflower mosaic virus (CaMV) 35S promoter were transformed into the irx9 and irx14 mutants. The transformation was performed independently twice and more than 64 independent transgenic lines for each construct were analyzed. The expression of OsGT43 genes in 8 representative transgenic lines for each construct was confirmed by reverse transcription PCR analysis of OsGT43 transcripts (). The irx9 mutation led to a reduction in stem mechanical strength down to 15 to 20% of that of the wild type due to a defect in secondary wall thickening (). Examination of transgenic irx9 lines expressing OsGT43 genes revealed that OsGT43A and OsGT43E were able to restore the stem mechanical strength to the wild-type level (). In contrast, expression of OsGT43H and OsGT43J in the irx9 mutant did not complement the stem mechanical strength. Consistent with the stem mechanical strength data, expression of OsGT43A and OsGT43E but not OsGT43H and OsGT43J rescued the collapsed vessel phenotypes caused by the irx9 mutation ().
Figure 7. Complementation of the irx9 and irx14 mutants by expression of OsGT43 genes. Rice GT43 cDNAs driven by the CaMV 35S promoter were introduced into irx9 or irx14 and the bottom parts of inflorescence stems of 10-wk-old transgenic overexpressors were examined for the stem breaking strength (B) and vessel morphology (C to N). Bar in (C) = 97 μm in (C) to (N). (A) Reverse transcription PCR analysis of expression of OsGT43 genes in 8 representative lines of transgenic irx9 or irx14 plants. The expression of the EF1α gene was used as an internal control. (B) Measurement of stem breaking strength of the wild type, irx9, irx14, and the mutants expressing OsGT43 genes. Each bar represents the breaking force of the inflorescence stem of individual plants. (C) to (H) The collapsed vessel phenotype (arrows) in irx9 (C) was rescued by expression of OsGT43A (D) and OsGT43E (E) but not OsGT43H (F) and OsGT43J (G). (I) to (N) The collapsed vessel phenotype (arrows) in irx14 (I) was rescued by expression of OsGT43J (M) but not OsGT43A (J), OsGT43E (K), and OsGT43H (L).
To determine whether the restored stem strength and vessel morphology were accompanied with the complementation of xylan defects, we examined xylan content and structure in irx9 expressing OsGT43 genes. Mature inflorescence stems of 10-wk-old plants from 8 independent transgenic lines were pooled for extraction of cell walls. Sugar composition analysis of cell walls from inflorescence stems revealed that although the xylose content in irx9 was only 52% of that of the wild type, expression of OsGT43A and OsGT43E restored the xylose content to about 80% of the wild-type level. In contrast, no restoration of xylose level was seen in the irx9 lines expressing OsGT43H and OsGT43J (). Similarly, the irx9 mutation caused a reduction of cellulose and lignin down to about 70% of the wild type, whereas expression of OsGT43A and OsGT43E but not OsGT43H and OsGT43J restored the levels of cellulose and lignin close to the wild-type level (). To analyze xylan structure, xylan was isolated from inflorescence stems and digested with endoxylanase to generate xylooligosaccharides, which were subsequently subjected to matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF-MS). The irx9 mutation caused a loss of GlcA side chains in xylan as evidenced by the absence of the signal peak corresponding to the GlcA-substituted Xyl4 (m/z 745) compared with the wild type (). A partial restoration of the signal peak for the GlcA-substituted Xyl4 was seen in the irx9 lines expressing OsGT43A () and OsGT43E () but not in those expressing OsGT43H () and OsGT43J (). The restoration of the GlcA side chains in the irx9 lines complemented with OsGT43A and OsGT43E was further verified by 1H nuclear magnetic resonance (NMR) spectroscopy (). It was shown that although the resonance peak characteristic of 1H GlcA at 5.31 ppm was absent in irx9 compared with the wild type, it was evident in irx9 complemented with OsGT43A and OsGT43E. In addition to the loss of GlcA side chains, the irx9 mutation caused a defect in xylan chain length that was shortened from an average of 93 in the wild type to 22 (), thereby resulting in a relative increase in the abundance of NMR signal peaks attributed to the xylan reducing end sequence, β-d-Xyl-(1→3)-α-l-Rha-(1→2)-α-d-GalA-(1→4)-d-Xyl (). Expression of OsGT43A and OsGT43E in irx9 led to a partial restoration of the xylan chain length () and concomitantly a significant reduction in the relative abundance of the xylan reducing end sequence ().
Table 1. Cell wall composition analysis of the irx9 and irx14 plants expressing rice GT43 genes
Figure 8. Measurement of cellulose and lignin amounts in irx9 and irx14 expressing OsGT43 genes. Cell walls were isolated from pooled mature inflorescence stems of 8 independent transgenic lines for each construct and used for measurement of the amounts of cellulose (A), guaiacyl lignin (B) and syringyl lignin (C). The data are mean ± SE of 2 separate assays. Note that expression of OsGT43A and OsGT43E in irx9 restored cellulose, guaiacyl lignin and syringyl lignin to the wild-type level and that expression of OsGT43J in irx14 restored cellulose, guaiacyl lignin and syringyl lignin to the wild-type level.
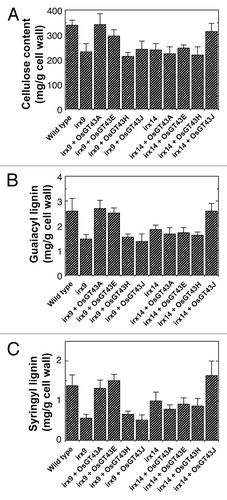
Figure 9. MALDI-TOF mass spectra of xylooligosaccharides generated by xylanase digestion of xylans from the wild type, irx9, irx14, and the mutants expressing OsGT43 genes. The ions at m/z 745 and 759 in the wild type (A and G) correspond to xylotetrasaccharides bearing a GlcA residue (X4G) or a methylated GlcA residue (X4M). (A) to (F) The missing ion at m/z 745 corresponding to X4G in irx9 (B) was partially restored by expression of OsGT43A (C) and OsGT43E (D) but not OsGT43H (E) and OsGT43J (F). (G) to (L) The missing ion at m/z 745 corresponding to X4G in irx14 (H) was restored by expression of OsGT43J (L) but not OsGT43A (I), OsGT43E (J), and OsGT43H (K).
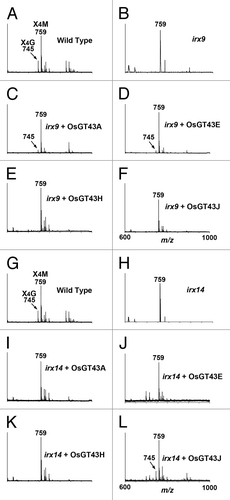
Figure 10.1H-NMR spectra of xylooligosaccharides generated by xylanase digestion of xylans from the wild type, irx9, irx14, and the mutants expressing OsGT43 genes. Resonances are labeled with the position of the assigned proton and the identity of the residue containing that proton. Note the restoration of the resonance of H1 of α-GlcA in irx9 complemented with OsGT43A and OsGT43E and in irx14 complemented with OsGT43J. The resonances of H1 of α-D-GalA, H1 of α-L-Rha, H1 of 3-linked β-D-Xyl, H4 of α-D-GalA, and H2 of α-L-Rha (arrow heads) are from the GX reducing end tetrasaccharide sequence. Note the relatively elevated resonance intensities of the GX reducing end tetrasaccharide sequence in irx 9 and irx14 compared with the wild type and the reduction in the intensity of these resonances in irx9 complemented with OsGT43A and OsGT43E and in irx14 complemented with OsGT43J.
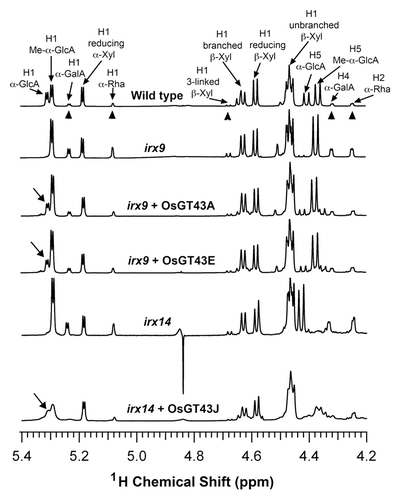
Table 2. Relative abundance of the reducing end sequence and the DP of xylan in the irx9 and irx14 plants expressing rice GT43 genes
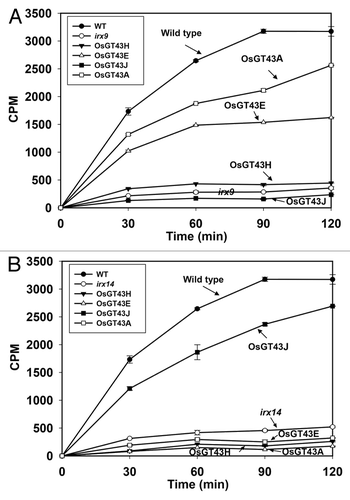
Consistent with the restored xylan content and structure, expression of OsGT43A and OsGT43E also rescued the defective xylosyltransferase activity conferred by the irx9 mutation (). While the xylan xylosyltransferase activity in irx9 microsomes was only 9% of the wild-type level, the activity in OsGT43A- and OsGT43E-complemented lines was restored to 52% and 78%, respectively, of the wild-type level. In contrast, expression of OsGT43H and OsGT43J did not result in any significant changes in the xylosyltransferase activity. Taken together, these results demonstrate that OsGT43A and OsGT43E are functional orthologs of IRX9 involved in the biosynthesis of the xylan backbone.
Figure 11. Time course of the xylosyltransferase activity in the microsomes from the wild type, irx9, irx14, and the mutants expressing OsGT43 genes. Microsomes were isolated from stems of 8 independent transgenic lines for each construct. The isolated microsomes were incubated with radiolabeled UDP-xylose and the Xyl4 acceptor, and the xylosyltransferase activity (CPM) was measured by the amount of radiolabeled xylosyl residues transferred onto the acceptor. The data are mean ± SE of 3 technical replicates. (A) Restoration of the xylosyltransferase activity in irx9 by expression of OsGT43A and OsGT43E but not OsGT43H and OsGT43J. (B) Restoration of the xylosyltransferase activity in irx14 by expression of OsGT43J but not OsGT43A, OsGT43E and OsGT43H.
OsGT43J but not OsGT43A, OsGT43E, and OsGT43H is a functional ortholog of Arabidopsis IRX14
Examination of transgenic irx14 plants expressing the 4 representative OsGT43 genes by comprehensive histological, biochemical, and structural analyses revealed that only OsGT43J complemented the irx14 mutant phenotypes, including the restoration of stem mechanical strength, vessel morphology, xylan content, celluose and lignin levels, GlcA side chains, xylan chain length, and xylosyltransferase activity (–; and ). Neither OsGT43A and OsGT43E nor OsGT43H was able to rescue the irx14 mutant phenotypes. These results demonstrate that OsGT43J is a functional ortholog of IRX14. Together with the complementation analysis in irx9, these results indicate that the rice GT43 family retained 2 functionally non-redundant groups of GT43 members. However, the functions of some of the rice GT43 members, such as OsGT43H, may have diverged from those of Arabidopsis.
Discussion
There exist 4 GT43 members in Arabidopsis, all of which have been demonstrated to be involved in xylan backbone biosynthesis. Previous genetic and biochemical analyses have shown that they form 2 functionally nonredundant groups, IRX9/IRX9 homolog and IRX14/IRX14 homolog, which act cooperatively in the elongation of the xylan backbone.Citation21-Citation23,Citation26-Citation28 The grass species such as rice apparently evolved to have an expansion of the GT43 family. Since expansion of a gene family is often accompanied by functional diversificationCitation36 and glycosyltransferase families are classified based on their amino acid sequence similarity rather than on their uses of common acceptors and/or donors,Citation37 one could not simply assume that all rice GT43 genes perform the same functions as those in Arabidopsis. Furthermore, it is currently unknown whether grass species evolved to retain 2 functionally nonredundant groups of GT43 genes for xylan biosynthesis because an early study of wheat protein complexes possessing xylan xylosyltransferase activity only identified an IRX14 homolog.Citation29 Therefore, it is critical to investigate whether the involvement of 2 functionally nonredundant groups of GT43 genes in xylan biosynthesis is evolutionarily conserved between dicots and grasses.
In this report, we studied the functions of rice GT43 members through complementation analysis in Arabidopsis irx9 and irx14 mutants. Phylogenetic analysis indicates that rice GT43 members are more diverged than those from Arabidopsis. In particular, there are 8 IRX9 homologs in rice, whereas only 2 members, IRX9 and IRX9 homolog, exist in Arabidopsis. Therefore, we selected 4 representative OsGT43 genes to carry out reciprocal complementation analysis in the Arabidopsis mutants. It was found that OsGT43A and OsGT43E, which are phylogenetically more related to IRX9, rescued the mutant phenotypes of irx9 but were unable to complement those of irx14. By contrast, OsGT43J, which is grouped closer to IRX14, rescued irx14 mutant phenotypes but was unable to complement those of irx9. These results demonstrate that OsGT43A and OsGT43E are orthologs of IRX9, and OsGT43J is an ortholog of IRX14. This finding provides the first line of genetic evidence indicating that rice evolved to retain 2 functionally nonredundant groups of GT43 genes involved in xylan backbone biosynthesis. The involvement of 2 functionally nonredundant groups of GT43 genes in xylan backbone biosynthesis is likely conserved in other grass species. A recent report showed that RNA interference suppression of an IRX9 homolog in wheat results in a decrease in arabinoxylan content.Citation38 Together with an early report of the presence of an IRX14 homolog in wheat protein complexes possessing xylan xylosyltransferase activity,Citation29 these reports indicate that homologs of both IRX9 and IRX14 are likely involved in xylan biosynthesis in wheat though it was still unknown whether wheat homologs of IRX9 and IRX14 are functionally nonredundant. Our finding further supports the hypothesis that 2 functionally nonredundant groups of GT43 xylosyltransferases are needed to overcome the steric problem for the transfer of xylosyl residues onto the growing xylan chain because one xylosyl residue is flipped nearly 180° relative to its neighboring residue.Citation26
In addition to the functional orthologs of IRX9 and IRX14, some rice GT43 members may have divergent functions. OsGT43H, which is phylogenetically close to I9H/IRX9L, could not complement the mutant phenotypes of either irx9 or irx14. One possibility is that OsGT43H might have evolved to have catalytic functions different from IRX9 and IRX14. It is common that glycosyltransferase members in the same GT families may perform different catalytic functions. For example, FRA8 and IRX10, both of which are phylogenetically closely related members of GT47 family, are involved in the biosynthesis of the xylan reducing end sequence and the xylan backbone, respectively.Citation23,Citation24 The other possibility is that although it could not complement irx9 and irx14 mutants due to its sequence divergence from the Arabidopsis GT43 protein, OsGT43H is still part of the xylan synthase complex in rice. It remains to be investigated whether the other OsGT43 member, OsGT43G, which is phylogenetically closely related to OsGT43H, is able to rescue the mutant phenotypes of irx9 or irx14.
It is interesting to note that although expression of OsGT43A and OsGT43B in irx9 and expression of OsGT43J in irx14 almost fully restored the stem strength, vessel morphology and xylose content to the wild-type levels, the chain length of xylan in the complemented plants is only restored to about 60% of that of the wild type. These results indicate that plants could tolerate a significant variation of xylan chain length and xylan with a 60% of wild-type xylan chain length is sufficient to accommodate the normal assembly of cell walls.
During the course of our investigation of the functions of rice GT43 proteins, a study reported that 2 other OsGT43 members, OsGT43C/OsIRX9 and OsGT43F/OsIRX9L, complemented the irx9 mutant phenotypes and OsGT43J/OsIRX14 complemented the irx14 mutant phenotypes.Citation39 However, it was stated that the defective xylan chain length in irx14, which is the primary defect of the mutant, was not complemented by examination of one presumably complemented transgenic line expressing OsGT43J/OsIRX14. Thus, the results from Chiniquy et al.Citation39 could not conclude that OsGT43J/OsIRX14 functionally complements the xylan defect exhibited by irx14. Our results demonstrated that OsGT43J/OsIRX14 was able to rescue the mutant phenotypes of irx14 including the defective xylan chain length (; ), which unequivocally establishes that OsGT43J is a functional ortholog of IRX14.
In summary, we show that rice stem microsomes possess both a xylan xylosyltransferase activity and a xylosidase-like activity, and the rice family GT43 retains 2 functionally non-redundant groups involved in xylan backbone biosynthesis. Although the xylan structure in rice differs from those in dicots by having no apparent reducing end sequence, abundant arabinosyl side chains, and ferulate as xylan cross-linking molecules, it appears that the biosynthetic machinery responsible for the xylan backbone elongation is conserved between rice and dicots, both of which involve 2 functionally nonredundant groups of GT43 members. Further investigation of the biochemical mechanism controlling xylan biosynthesis in rice could provide knowledge basis for genetic modification of grass crops, such as switchgrass and Miscanthus, better suited for biofuel production.
Materials and Methods
Cell wall preparation
Rice (Oryza sativa Japonica Teipei 309) stems and Arabidopsis (Arabidopsis thaliana) inflorescence stems were used for cell wall isolation according to Zhong et al.Citation35 Cell walls were extracted for xylan with 1N KOH, and the solubilized xylan was digested with β-xylanase M6 (Megazyme) to generate xylooligosaccharide for matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) and nuclear magnetic resonance (NMR) spectroscopy.
1H-NMR spectroscopy
NMR spectra of rice and Arabidopsis xylooligosaccharides were acquired at 20 °C on a 600 MHz spectrometer (599.7 MHz, 1H) using a 3 mm cryogenic triple resonance probe.Citation40 All NMR samples were prepared with 100% D2O. For all experiments, 128 transients were collected using a spectral width of 6,000 Hz and an acquisition time of 5 s. The residual water resonance was suppressed by a 1 s presaturation pulse at a field strength of 40 Hz. All spectra were processed with 0.2-Hz apodization followed by zero-filling to 128 k points. The 1H NMR assignments were done by comparing them to the NMR spectral data for xylan structure.Citation23,Citation31,Citation32,Citation35
Assay of xylosyltransferase activity
Microsomes were isolated from the stems of rice and Arabidopsis following the procedure of Kuroyama and TsumurayCitation33 and stored at -80 °C until used. For assay of the xylosyltransferase activity using the radiolabeling method, microsomes (100 μg) were incubated with the reaction mixture containing 50 mM HEPES-KOH, pH 6.8, 5 mM MnCl2, 1 mM dithiothritol, 0.5% Triton X-100, 0.1 mM cold UDP-Xyl (CarboSource Service), 0.2 μg/μl Xyl4 (Megazyme) and UDP-[14C]Xyl (0.1 μCi; American Radiolabeled Chemical) at 21 °C for 30 min. The reaction products were separated from UDP-[14C]Xyl by paper chromatography according to Ishikawa et al.Citation41 and counted for the amount of radioactivity with a PerkinElmer scintillation counter. For assay of the xylosyltransferase activity using fluorescent anthranilic acid (AA)-labeled xylooligomer acceptors, microsomes (100 μg) were incubated with the reaction mixture containing 50 mM HEPES-KOH, pH 6.8, 5 mM MnCl2, 1 mM DTT, 0.5% Triton X-100, 0.1 mM cold UDP-Xyl, 0.5 mM Xyln-AA at 21 °C for 30 min. The reaction products were analyzed by reversed-phase HPLC analysis. HPLC was performed with a PerkinElmer series 200 LC system and a PerkinElmer series 200a fluorescence detector (Ex320nm, Em420nm). The incorporated Xyln-AA products were separated using a Kinetex C18 (2.6 μm) column (100 mm long, 4.6 mm i.d.) (Phenomenex) according to Lee et al.Citation22
Xylosidase digestion of xylooliogmers
The rice microsome-catalyzed reaction products using the Xyl6-AA acceptor were incubated with β-(1,4)-xylosidase (0.005 U; Sigma) for 1 h at 37 °C. The reaction was terminated with 0.1 M acetic acid and subjected to reverse-phase HPLC analysis.
Subcellular localization
The subcellular localization of OsGT43 proteins was performed by co-transfecting yellow fluorescent protein (YFP)-tagged fusion proteins and the cyan fluorescent protein (CFP)-tagged Golgi marker (FRA8) into Arabidopsis protoplasts.Citation42 The full-length OsGT43 cDNAs were fused in frame with the YFP cDNA and ligated between the CaMV 35S promoter and the nopaline synthase terminator in pBI221 (Clontech). After transfection, protoplasts were incubated for 16 h and fluorescence signals in transfected protoplasts were visualized using a Leica TCs SP2 spectral confocal microscope (Leica Microsystems).
Complementation of the irx9 and irx14 mutants
The full-length OsGT43 cDNAs driven by the CaMV 35S promoter were cloned into the pGPTV binary vector containing hygromycin resistant selection marker geneCitation43 to create OsGT43 expression constructs, and the cloned OsGT43 cDNA sequences in the expression constructs were verified by sequencing. The OsGT43 expression constructs were introduced into the Arabidopsis irx9 or irx14 mutant by the agrobacterium-mediated transformation. Transgenic plants were selected on hygromycin, and the expression of OsGT43 genes in rosette leaves of transgenic plants was verified using reverse-transcription PCR. Inflorescence stems from the first generation of complemented transgenic plants were used for morphological and chemical analyses of phenotypes. Basal parts of the main inflorescence of 10-wk-old plants were measured for the breaking force using a digital force/length tester.Citation44 The breaking force was calculated as the force needed to break apart a stem segment. Basal parts of the main inflorescence of 10-wk-old plants were also fixed for light microscopy.Citation45 For each construct, at least 64 transgenic plants were generated and examined.
Cell wall analysis
Cell wall sugars (as alditol acetates) were assayed following the procedure described by Hoebler et al.Citation46 The alditol acetates of the hydrolyzed cell wall sugars were analyzed on a PerkinElmer Clarus 500 gas-liquid chromatograph instrument equipped with a 30 min x 0.25 mm (i.d.) silica capillary column DB 225 (Alltech Assoc.). Cellulose content was assayed with the anthrone reagent according to Updegraff.Citation47 Lignin composition was determined as described.Citation48,Citation49 Cell walls were hydrolyzed in 4 N NaOH and the released lignin monomers were extracted into diethylether and vacuum dried. The residue was dissolved in pyridine and N,O-bis(trimethylsilyl)trifluoroacetamide and analyzed for phenolics by gas chromatography. Phenolic compounds were identified by comparison of their mass spectra with those of the authentic compounds. Guaiacyl lignin unit is the sum of vanillin, acetovanillin, and vanillic acid. Syringyl lignin unit is the sum of syringaldehyde, acetosyringaldehyde, and syringic acid.
MALDI-TOF MS
Xylooligosaccharides released from xylanase digestion of KOH-extracted xylan were analyzed using a MALDI-TOF mass spectrometer operated in the positive-ion mode with an accelerating voltage of 30 kV, an extractor voltage of 9 kV, and a source pressure of approximately 8 x 10−7 torr. The aqueous sample was mixed (1:1, v/v) with the MALDI matrix (0.2 M 2,5-dihydroxbenzoic acid and 0.06 M 1-hydroxyisoquinoline in 50% acetonitrile) and dried on the stainless steel target plate. Spectra are the average of 100 laser shots.
Accession numbers
The locus identifiers for the rice GT43 genes are OsGT43A (LOC_Os05 g03174; Os05 g0123100), OsGT43B (LOC_Os03 g17850; Os03 g0287800), OsGT43C/OsIRX9 (LOC_Os07 g49370; Os07 g0694400), OsGT43D (LOC_Os01 g06450; Os01 g0157700), OsGT43E (LOC_Os05 g48600; Os05 g0559600), OsGT43F/OsIRX9L (LOC_01 g48440; Os01 g0675500), OsGT43G (LOC_Os10 g13810; Os10 g0205300), OsGT43H (LOC_Os04 g01280; Os04 g0103100), OsGT43I (LOC_Os04 g55670; Os04 g0650300), and OsGT43J/OsIRX14 (LOC_Os06 g47340; Os06 g0687900). The Arabidopsis Genome Initiative locus identifiers for the Arabidopsis GT43 genes are IRX9 (At2g37090), I9H/IRX9L (At1g27600), IRX14 (At4g36890), and I14H/IRX14L (At5g67230). The GenBank accession numbers of the rice GT43 genes are KJ206898 for OsGT43A, KJ206899 for OsGT43B, KJ206900 for OsGT43C, KJ206901 for OsGT43D, KJ206902 for OsGT43E, KJ206903 for OsGT43F, KJ206904 for OsGT43G, KJ206905 for OsGT43H, KJ206906 for OsGT43I, and KJ206907 for OsGT43J.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was funded by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy (DE-FG02–03ER15415).
References
- Carroll A, Somerville C. Cellulosic biofuels. Annu Rev Plant Biol 2009; 60:165 - 82; http://dx.doi.org/10.1146/annurev.arplant.043008.092125; PMID: 19014348
- McCann MC, Carpita NC. Designing the deconstruction of plant cell walls. Curr Opin Plant Biol 2008; 11:314 - 20; http://dx.doi.org/10.1016/j.pbi.2008.04.001; PMID: 18486537
- Ebringerová A, Heinze T. Xylan and xylan derivatives-biopolymers with valuable properties, 1. Naturally occurring xylans structures, isolation procedures and properties. Macromol Rapid Commun 2000; 21:542 - 56; http://dx.doi.org/10.1002/1521-3927(20000601)21:9<542::AID-MARC542>3.0.CO;2-7
- Scheller HV, Ulvskov P. Hemicelluloses. Annu Rev Plant Biol 2010; 61:263 - 89; http://dx.doi.org/10.1146/annurev-arplant-042809-112315; PMID: 20192742
- Oikawa A, Joshi HJ, Rennie EA, Ebert B, Manisseri C, Heazlewood JL, Scheller HV. An integrative approach to the identification of Arabidopsis and rice genes involved in xylan and secondary wall development. PLoS One 2010; 5:e15481; http://dx.doi.org/10.1371/journal.pone.0015481; PMID: 21124849
- Bosch M, Mayer CD, Cookson A, Donnison IS. Identification of genes involved in cell wall biogenesis in grasses by differential gene expression profiling of elongating and non-elongating maize internodes. J Exp Bot 2011; 62:3545 - 61; http://dx.doi.org/10.1093/jxb/err045; PMID: 21402660
- Pellny TK, Lovegrove A, Freeman J, Tosi P, Love CG, Knox JP, Shewry PR, Mitchell RA. Cell walls of developing wheat starchy endosperm: comparison of composition and RNA-Seq transcriptome. Plant Physiol 2012; 158:612 - 27; http://dx.doi.org/10.1104/pp.111.189191; PMID: 22123899
- Anders N, Wilkinson MD, Lovegrove A, Freeman J, Tryfona T, Pellny TK, Weimar T, Mortimer JC, Stott K, Baker JM, et al. Glycosyl transferases in family 61 mediate arabinofuranosyl transfer onto xylan in grasses. Proc Natl Acad Sci U S A 2012; 109:989 - 93; http://dx.doi.org/10.1073/pnas.1115858109; PMID: 22215597
- Chiniquy D, Sharma V, Schultink A, Baidoo EE, Rautengarten C, Cheng K, Carroll A, Ulvskov P, Harholt J, Keasling JD, et al. XAX1 from glycosyltransferase family 61 mediates xylosyltransfer to rice xylan. Proc Natl Acad Sci U S A 2012; 109:17117 - 22; http://dx.doi.org/10.1073/pnas.1202079109; PMID: 23027943
- Piston F, Uauy C, Fu L, Langston J, Labavitch J, Dubcovsky J. Down-regulation of four putative arabinoxylan feruloyl transferase genes from family PF02458 reduces ester-linked ferulate content in rice cell walls. Planta 2010; 231:677 - 91; http://dx.doi.org/10.1007/s00425-009-1077-1; PMID: 20012086
- Bartley LE, Peck ML, Kim SR, Ebert B, Manisseri C, Chiniquy DM, Sykes R, Gao L, Rautengarten C, Vega-Sánchez ME, et al. Overexpression of a BAHD acyltransferase, OsAt10, alters rice cell wall hydroxycinnamic acid content and saccharification. Plant Physiol 2013; 161:1615 - 33; http://dx.doi.org/10.1104/pp.112.208694; PMID: 23391577
- Mortimer JC, Miles GP, Brown DM, Zhang Z, Segura MP, Weimar T, Yu X, Seffen KA, Stephens E, Turner SR, et al. Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. Proc Natl Acad Sci U S A 2010; 107:17409 - 14; http://dx.doi.org/10.1073/pnas.1005456107; PMID: 20852069
- Lee C, Teng Q, Zhong R, Ye Z-H. Arabidopsis GUX proteins are glucuronyltransferases responsible for the addition of glucuronic acid side chains onto xylan. Plant Cell Physiol 2012; 53:1204 - 16; http://dx.doi.org/10.1093/pcp/pcs064; PMID: 22537759
- Rennie EA, Hansen SF, Baidoo EE, Hadi MZ, Keasling JD, Scheller HV. Three members of the Arabidopsis glycosyltransferase family 8 are xylan glucuronosyltransferases. Plant Physiol 2012; 159:1408 - 17; http://dx.doi.org/10.1104/pp.112.200964; PMID: 22706449
- Lee C, Teng Q, Zhong R, Yuan Y, Haghighat M, Ye Z-H. Three Arabidopsis DUF579 domain-containing GXM proteins are methyltransferases catalyzing 4-o-methylation of glucuronic acid on xylan. Plant Cell Physiol 2012; 53:1934 - 49; http://dx.doi.org/10.1093/pcp/pcs138; PMID: 23045523
- Urbanowicz BR, Peña MJ, Ratnaparkhe S, Avci U, Backe J, Steet HF, Foston M, Li H, O’Neill MA, Ragauskas AJ, et al. 4-O-methylation of glucuronic acid in Arabidopsis glucuronoxylan is catalyzed by a domain of unknown function family 579 protein. Proc Natl Acad Sci U S A 2012; 109:14253 - 8; http://dx.doi.org/10.1073/pnas.1208097109; PMID: 22893684
- Lee C, Teng Q, Zhong R, Ye Z-H. The four Arabidopsis reduced wall acetylation genes are expressed in secondary wall-containing cells and required for the acetylation of xylan. Plant Cell Physiol 2011; 52:1289 - 301; http://dx.doi.org/10.1093/pcp/pcr075; PMID: 21673009
- Yuan Y, Teng Q, Zhong R, Ye Z-H. The Arabidopsis DUF231 domain-containing protein ESK1 mediates 2-O- and 3-O-acetylation of xylosyl residues in xylan. Plant Cell Physiol 2013; 54:1186 - 99; http://dx.doi.org/10.1093/pcp/pct070; PMID: 23659919
- Xiong G, Cheng K, Pauly M. Xylan O-acetylation impacts xylem development and enzymatic recalcitrance as indicated by the Arabidopsis mutant tbl29.. Mol Plant 2013; 6:1373 - 5; http://dx.doi.org/10.1093/mp/sst014; PMID: 23340742
- Lerouxel O, Cavalier DM, Liepman AH, Keegstra K. Biosynthesis of plant cell wall polysaccharides - a complex process. Curr Opin Plant Biol 2006; 9:621 - 30; http://dx.doi.org/10.1016/j.pbi.2006.09.009; PMID: 17011813
- Brown DM, Goubet F, Wong VW, Goodacre R, Stephens E, Dupree P, Turner SR. Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J 2007; 52:1154 - 68; http://dx.doi.org/10.1111/j.1365-313X.2007.03307.x; PMID: 17944810
- Lee C, O’Neill MA, Tsumuraya Y, Darvill AG, Ye Z-H. The irregular xylem9 mutant is deficient in xylan xylosyltransferase activity. Plant Cell Physiol 2007; 48:1624 - 34; http://dx.doi.org/10.1093/pcp/pcm135; PMID: 17938130
- Peña MJ, Zhong R, Zhou G-K, Richardson EA, O’Neill MA, Darvill AG, York WS, Ye Z-H. Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 2007; 19:549 - 63; http://dx.doi.org/10.1105/tpc.106.049320; PMID: 17322407
- Brown DM, Zhang Z, Stephens E, Dupree P, Turner SR. Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis.. Plant J 2009; 57:732 - 46; http://dx.doi.org/10.1111/j.1365-313X.2008.03729.x; PMID: 18980662
- Wu AM, Rihouey C, Seveno M, Hörnblad E, Singh SK, Matsunaga T, Ishii T, Lerouge P, Marchant A. The Arabidopsis IRX10 and IRX10-LIKE glycosyltransferases are critical for glucuronoxylan biosynthesis during secondary cell wall formation. Plant J 2009; 57:718 - 31; http://dx.doi.org/10.1111/j.1365-313X.2008.03724.x; PMID: 18980649
- Lee C, Teng Q, Huang W, Zhong R, Ye Z-H. The Arabidopsis family GT43 glycosyltransferases form two functionally nonredundant groups essential for the elongation of glucuronoxylan backbone. Plant Physiol 2010; 153:526 - 41; http://dx.doi.org/10.1104/pp.110.155309; PMID: 20335400
- Wu AM, Hörnblad E, Voxeur A, Gerber L, Rihouey C, Lerouge P, Marchant A. Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L pairs of glycosyltransferase genes reveals critical contributions to biosynthesis of the hemicellulose glucuronoxylan. Plant Physiol 2010; 153:542 - 54; http://dx.doi.org/10.1104/pp.110.154971; PMID: 20424005
- Lee C, Zhong R, Ye Z-H. Arabidopsis family GT43 members are xylan xylosyltransferases required for the elongation of the xylan backbone. Plant Cell Physiol 2012; 53:135 - 43; http://dx.doi.org/10.1093/pcp/pcr158; PMID: 22080591
- Zeng W, Jiang N, Nadella R, Killen TL, Nadella V, Faik A. A glucurono(arabino)xylan synthase complex from wheat contains members of the GT43, GT47, and GT75 families and functions cooperatively. Plant Physiol 2010; 154:78 - 97; http://dx.doi.org/10.1104/pp.110.159749; PMID: 20631319
- Chen X, Vega-Sánchez ME, Verhertbruggen Y, Chiniquy D, Canlas PE, Fagerström A, Prak L, Christensen U, Oikawa A, Chern M, et al. Inactivation of OsIRX10 leads to decreased xylan content in rice culm cell walls and improved biomass saccharification. Mol Plant 2013; 6:570 - 3; http://dx.doi.org/10.1093/mp/sss135; PMID: 23180670
- Hoffmann RA, Geijtenbeek T, Kamerling JP, Vliegenthart JF. 1H-N.m.r. study of enzymically generated wheat-endosperm arabinoxylan oligosaccharides: structures of hepta- to tetradeca-saccharides containing two or three branched xylose residues. Carbohydr Res 1992; 223:19 - 44; http://dx.doi.org/10.1016/0008-6215(92)80003-J; PMID: 1596918
- Verbruggen MA, Spronk BA, Schols HA, Beldman G, Voragen AG, Thomas JR, Kamerling JP, Vliegenthart JF. Structures of enzymically derived oligosaccharides from sorghum glucuronoarabinoxylan. Carbohydr Res 1998; 306:265 - 74; http://dx.doi.org/10.1016/S0008-6215(97)10064-7; PMID: 9691450
- Kuroyama H, Tsumuraya Y. A xylosyltransferase that synthesizes β-(1-->4)-xylans in wheat (Triticum aestivum L.) seedlings. Planta 2001; 213:231 - 40; http://dx.doi.org/10.1007/s004250000499; PMID: 11469588
- Lee C, Zhong R, Ye Z-H. Biochemical characterization of xylan xylosyltransferases involved in wood formation in poplar. Plant Signal Behav 2012; 7:332 - 7; http://dx.doi.org/10.4161/psb.19269; PMID: 22476457
- Zhong R, Peña MJ, Zhou G-K, Nairn CJ, Wood-Jones A, Richardson EA, Morrison WH 3rd, Darvill AG, York WS, Ye Z-H. Arabidopsis fragile fiber8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell 2005; 17:3390 - 408; http://dx.doi.org/10.1105/tpc.105.035501; PMID: 16272433
- Bellieny-Rabelo D, Oliveira AE, Venancio TM. Impact of whole-genome and tandem duplications in the expansion and functional diversification of the F-box family in legumes (Fabaceae). PLoS One 2013; 8:e55127; http://dx.doi.org/10.1371/journal.pone.0055127; PMID: 23390519
- Campbell JA, Davies GJ, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J 1997; 326:929 - 39; PMID: 9334165
- Lovegrove A, Wilkinson MD, Freeman J, Pellny TK, Tosi P, Saulnier L, Shewry PR, Mitchell RA. RNA interference suppression of genes in glycosyl transferase families 43 and 47 in wheat starchy endosperm causes large decreases in arabinoxylan content. Plant Physiol 2013; 163:95 - 107; http://dx.doi.org/10.1104/pp.113.222653; PMID: 23878080
- Chiniquy D, Varanasi P, Oh T, Harholt J, Katnelson J, Singh S, Auer M, Simmons B, Adams PD, Scheller HV, et al. Three novel rice genes closely related to the Arabidopsis IRX9, IRX9L, and IRX14 genes and their roles in xylan biosynthesis. Front Plant Sci 2013; 4:83; http://dx.doi.org/10.3389/fpls.2013.00083; PMID: 23596448
- Teng Q, Ekman DR, Huang W, Collette TW. Push-through direct injection NMR: an optimized automation method applied to metabolomics. Analyst 2012; 137:2226 - 32; http://dx.doi.org/10.1039/c2an16251b; PMID: 22434060
- Ishikawa M, Kuroyama H, Takeuchi Y, Tsumuraya Y. Characterization of pectin methyltransferase from soybean hypocotyls. Planta 2000; 210:782 - 91; http://dx.doi.org/10.1007/s004250050680; PMID: 10805450
- Sheen J. Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 2001; 127:1466 - 75; http://dx.doi.org/10.1104/pp.010820; PMID: 11743090
- Becker D, Kemper E, Schell J, Masterson R. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 1992; 20:1195 - 7; http://dx.doi.org/10.1007/BF00028908; PMID: 1463855
- Zhong R, Taylor JJ, Ye Z-H. Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell 1997; 9:2159 - 70; PMID: 9437861
- Burk DH, Zhong R, Morrison WHIII, Ye Z-H. Disruption of cortical microtubules by overexpression of green fluorescent protein-tagged α-tubulin 6 causes a marked reduction in cell wall synthesis. J Integr Plant Biol 2006; 48:85 - 98; http://dx.doi.org/10.1111/j.1744-7909.2006.00202.x
- Hoebler C, Barry JL, David A, Delort-Laval J. Rapid acid-hydrolysis of plant cell wall polysaccharides and simplified quantitative determination of their neutral monosaccharides by gas-liquid chromatography. J Agric Food Chem 1989; 37:360 - 7; http://dx.doi.org/10.1021/jf00086a020
- Updegraff DM. Semimicro determination of cellulose in biological materials. Anal Biochem 1969; 32:420 - 4; http://dx.doi.org/10.1016/S0003-2697(69)80009-6; PMID: 5361396
- Akin DE, Morrison WH, Himmelsbach DS. Characterization of digestion residues of alfalfa and orchardgrass leaves by microscopic, spectroscopic and chemical analysis. J Sci Food Agric 1993; 63:339 - 47; http://dx.doi.org/10.1002/jsfa.2740630312
- Zhong R, Iii WH, Negrel J, Ye Z-H. Dual methylation pathways in lignin biosynthesis. Plant Cell 1998; 10:2033 - 46; PMID: 9836743
- Li M, Xu W, Yang W, Kong Z, Xue Y. Genome-wide gene expression profiling reveals conserved and novel molecular functions of the stigma in rice. Plant Physiol 2007; 144:1797 - 812; http://dx.doi.org/10.1104/pp.107.101600; PMID: 17556504
