Abstract
Brassinosteroids (BRs) play important roles in plant growth, development and responses to environmental cues. BRs signal through plasma membrane-localized receptors to control BES1/BZR1 family transcription factors, which mediate the expression of thousands of genes. BRs activate and repress approximately equal numbers of genes. BES1/BZR1 interact with other transcription factors, histone-modifying enzymes, and transcription elongation factors to activate BR-induced genes. However, the mechanisms by which BES1/BZR1 mediate the BR-repressed gene expression are not well understood. Recent studies revealed that 2 BR-repressed transcription factors, MYBL2 and HAT1, cooperate with BES1 to downregulate BR-repressed genes expression. Moreover, BIN2 kinase, a well-established negative regulator in the BR signaling pathway, phosphorylates MYBL2 and HAT1. While BIN2 phosphorylates and destabilizes BES1/BZR1, BIN2 phosphorylated MYBL2 and HAT1 appear to be stabilized. These results not only extended our understanding of BR-repressed gene expression, but also revealed multiple inputs of BR signaling into BR transcriptional networks.
Plant steroid hormones, Brassinosteroids (BRs), play prominent roles in plant growth and development, including cell elongation, vascular differentiation, reproduction, senescence and responses to both biotic and abiotic stresses. BRs signal through receptor kinase BRI1 (BRASSINOSTEROID-INSENSITIVE1), co-receptor BAK1 (BRI1-ASSOCIATED RECEPTOR KINASE 1), and several other signaling components to control BES1 (BRI1 EMS SUPRESSOR 1) and BZR1 (BRASSINAZOLE RESISTANT 1) family transcription factors.Citation1 Global gene expression studies demonstrated that BRs can regulate up to 4000–5000 genes at different growth stages, with about half induced and half-repressed by BRs.Citation2 ChIP-Chip analyses indicated that BES1 and BZR1 transcription factors likely account for a majority of the BR-regulated gene expression, as BES1 and BZR1 have thousands of target genes and many of them are regulated by BRs.Citation3,Citation4
Significant progress has been made in understanding how BES1 and BZR1 regulate gene expression, especially on how they activate gene expression.Citation2,Citation5 BES1 can interact with other transcription factors such as BIM1, MYB30, PIF4, DELLA repressors to amplify BR signal, to integrate with other signaling pathways such as light and gibberellin in the regulation of plant growth.Citation6-Citation11 In addition, BES1 was shown to recruit histone modifying enzymes and a transcription elongation factor to modulate target gene expression.Citation12,Citation13 The mechanisms by which BES1/BZR1 represses thousands of genes have just begun to be revealed.
BIN2 (BRASSOSTERROIS INSENSITIVE 2), a member of GSK3 like kinase, is a negative regulator in BR signaling pathway.Citation14 BIN2 phosphorylates BES1/BZR1 family transcription factors and inhibits their functions through several mechanisms, including protein degradation, reducing DNA binding, and cytoplasmic retention.Citation5 BIN2 can also phosphorylates and modulates many other signaling components involved in BR or other signaling pathways. BIN2 phosphorylates ARF2 (AUXIN RESPONSE FACTOR 2), YODA MAP kinase kinase kinase, MKK4 (MAP KINASE KINASE 4), SPCH (SPEECHLESS), and DLT (DWARF AND LOW-TILLERING) to inhibit their functions in regulating growth and stomata development.Citation15-Citation19 While BIN2 phosphorylates and inhibits the functions of most of its known substrates, recent studies on HAT1 and MYBL2 demonstrated that BIN2 phosphorylation can have different outcomes.
MYBL2 (MYELOBLASTOSIS-LIKE 2) is a small MYB protein that has only one R3-MYB (SANT) domain. MYBL2 interacts with a basic helix-loop-helix transcription factor, to repress anthocyanin biosynthesis gene expression.Citation20,Citation21 MYBL2 was identified as BES1 and BZR1 direct target gene and is repressed by BRs.Citation22 MYBL2 is a positive regulator of BR-regulated plant growth as loss-of-function mybl2 mutant enhances the dwarf phenotype of a weak allele of bri1 and suppresses the constitutive BR-response mutant bes1-D. It was shown that the SANT domain of MYBL2 is involved in interaction with BES1 and such interaction appears to cooperatively repress BR-repressed gene expression. Furthermore, BIN2 phosphorylates MYBL2, which stabilized MYBL2 protein.Citation22
HAT1 (HOMEOBOX ARABIDOPSIS THALIANA 1) belongs to homeodomain-leucine zipper (HD-ZIP) family II, which has 9 members in Arabidopsis.Citation23,Citation24 HD-ZIP II members can bind to partially inverted repeats such as CAAT(A/T)ATTG, CAAT(C/G)ATTG, or slightly changed version TAAT(C/T)ATTA.Citation23 The HD-ZIP II family members are involved in various biological processes such as shade avoidance, meristem determination, leaf polarity, gynoecium and fruit development, auxin and other hormone responses.Citation23,Citation25-Citation29 The fact that HAT1 and its close homologs, HAT2, HAT3, and HAT4 (ATHB-2), are BES1/BZR1 direct target genes,Citation3,Citation4 prompt us to examine the function of the family transcription factors in BR responses. HAT1 and HAT3 are positive regulators for BR-regulated growth as double mutants of hat1 hat3 have reduced BR response in hypocotyl elongation assays and overexpression of HAT1 leads to increased stem elongation at the early stage of vegetative growth.Citation30 We further found that HAT1 binds to a conserved DNA binding site (TAATAATTA), interacts with BES1 bound to BRRE site to cooperatively inhibit BR-repressed gene expression. The results suggest that HAT1 function as a co-repressor of BES1, providing a mechanism for BES1 to repress gene expression.Citation30
Interestingly, HAT1 has more than 20 putative phosphorylation sites for BIN2 kinase and is phosphorylated by BIN2 kinase in vitro and likely in vivo as well.Citation30 Unlike BIN2 phosphorylation of BES1/BZR1, BIN2 phosphorylation appears to stabilize HAT1 protein,Citation30 which is similar to BIN2 phosphorylation of MYBL2.Citation22 What is the biological implication of BIN2 regulation of these transcriptional co-repressors? First, since MYBL2 and HAT1 are positive regulators of BR-regulated growth (by acting as co-repressors of BES1 in inhibiting BR-repressed gene expression), BIN2 phosphorylation / stabilization of these co-repressors when there are no BRs or non-phosphorylation / destabilization in the presence of BR provide mechanisms for feedback control by BRs. In other words, these transcription factors can function as “buffers” to balance BR-repressed gene expression under various BR concentrations.Citation22 Under low BRs, active BIN2 phosphorylates and stabilizes HAT1/MYBL2, which can cooperate with and compensate for reduced level of BES1 in maintaining BR-repressed gene expression. Under high BR concentrations in which BIN2 is inhibited, reduced levels of HAT1/MYBL2 likely balance out increased BES1 in the regulation of BR-repressed gene expression. Under optimal condition (moderate level of BRs), unphosphorylated BES1 can interact with phosphorylated MYBL2; and phosphorylated HAT1 bound to its binding site can cooperate with BES1 to regulate BR-repressed genes expression (). Thus the differential regulation of BES1/BZR1 (and other transcription factors such as SPCH and DLT) and MYBL2/HAT1 by BIN2 kinase provides a dynamic system to achieve stable growth under various conditions.
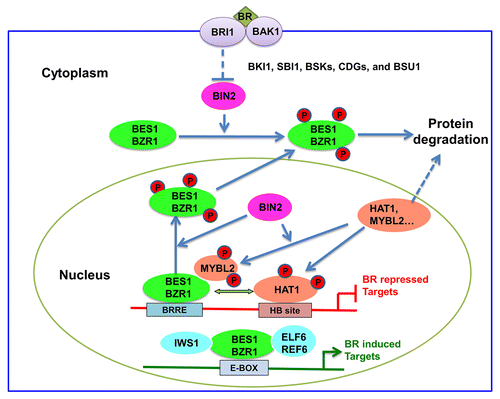
Figure 1. A proposed model for MYBL2/HAT1 functions in the BR pathway. The plants respond to BR signaling and maintain balanced levels of BES1/BZR1, MYBL2 and HAT1 to regulate BR-repressed genes. BIN2 phosphorylation can inhibit BES1/BZR1 functions but active HAT1/MYBL2 functions. Dephosphorylated BES1/BZR1 interact with phosphorylated MYBL2 and phosphorylated HAT1 to regulate BR-repressed genes expression. While MYBL2 does not bind DNA directly, BES1/BZR1 and HAT1 bind to BRRE and HB (Homeobox Binding) site, respectively, in BR-repressed gene promoters. On BR-induced gene promoters, BES1/BZR1 bound to E-box interact with other transcription factors (e.g., BIM1, MYB30, and PIF4, not shown) or regulators such as ELF6/REFCitation6 and IWS1 to activate gene expression.
The identification of MYBL2 and HAT1 as BES1 co-repressors and their unique regulation by BIN2 phosphorylation provide a framework to understand how BRs repress a large number of genes and how the process is regulated by BR signaling. Several questions need to be addressed. First, do these BR-repressed co-repressors account for a subset of BR-repressed genes and specific aspects of BR responses? Global gene expression studies and identification of their direct target genes can help answer the question. It’s very interesting that BIN2 phosphorylation has opposite effects on HAT1/MYBL2 (stabilization) and on BES1/BZR1/DLT/SPCH (destabilization). What are the molecular mechanisms underlying these differential regulations? Detailed biochemical and molecular genetic studies can help address the questions. Finally, BES1 and BZR1 directly regulate about 200 other transcription factors (termed BES1/BZR1 Targeted Transcription Factors, or BTFs), half of which are repressed by BRs and BES1/BZR1.Citation2 Do some of these BTFs function similarly to HAT1/MYBL2 in BR-repressed gene expression and are similarly (or differently) regulated by BIN2? Functional characterization of these BTFs can help address the questions. These studies should generate a more complete picture of the complex transcriptional network and the integration of BR signaling and BR-regulated gene expression.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by United States National Science Foundation (NSF) grant (IOS-1257631) and Plant Science Institute (to YY), Doctoral Foundation of the Ministry of Education (20110181110059 and 20120181130008), and Sichuan Nature Science Foundation (2010JQ0080).
References
- Zhu JY, Sae-Seaw J, Wang ZY. Brassinosteroid signalling. Development 2013; 140:1615 - 20; http://dx.doi.org/10.1242/dev.060590; PMID: 23533170
- Guo H, Li L, Aluru M, Aluru S, Yin Y. Mechanisms and networks for brassinosteroid regulated gene expression. Curr Opin Plant Biol 2013; 16:545 - 53; http://dx.doi.org/10.1016/j.pbi.2013.08.002; PMID: 23993372
- Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, Guo H, Anderson S, Aluru S, Liu P, et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J 2011; 65:634 - 46; http://dx.doi.org/10.1111/j.1365-313X.2010.04449.x; PMID: 21214652
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 2010; 19:765 - 77; http://dx.doi.org/10.1016/j.devcel.2010.10.010; PMID: 21074725
- Li J. Regulation of the nuclear activities of brassinosteroid signaling. Curr Opin Plant Biol 2010; 13:540 - 7; http://dx.doi.org/10.1016/j.pbi.2010.08.007; PMID: 20851039
- Oh E, Zhu JY, Wang ZY. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 2012; 14:802 - 9; http://dx.doi.org/10.1038/ncb2545; PMID: 22820378
- Li QF, Wang C, Jiang L, Li S, Sun SS, He JX. An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci Signal 2012; 5:ra72; http://dx.doi.org/10.1126/scisignal.2002908; PMID: 23033541
- Gallego-Bartolomé J, Minguet EG, Grau-Enguix F, Abbas M, Locascio A, Thomas SG, Alabadí D, Blázquez MA. Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc Natl Acad Sci U S A 2012; 109:13446 - 51; http://dx.doi.org/10.1073/pnas.1119992109; PMID: 22847438
- Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun TP, Wang ZY. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol 2012; 14:810 - 7; http://dx.doi.org/10.1038/ncb2546; PMID: 22820377
- Li L, Yu X, Thompson A, Guo M, Yoshida S, Asami T, Chory J, Yin Y. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J 2009; 58:275 - 86; http://dx.doi.org/10.1111/j.1365-313X.2008.03778.x; PMID: 19170933
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 2005; 120:249 - 59; http://dx.doi.org/10.1016/j.cell.2004.11.044; PMID: 15680330
- Li L, Ye H, Guo H, Yin Y. Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc Natl Acad Sci U S A 2010; 107:3918 - 23; http://dx.doi.org/10.1073/pnas.0909198107; PMID: 20139304
- Yu X, Li L, Li L, Guo M, Chory J, Yin Y. Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc Natl Acad Sci U S A 2008; 105:7618 - 23; http://dx.doi.org/10.1073/pnas.0802254105; PMID: 18467490
- Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 2002; 295:1299 - 301; http://dx.doi.org/10.1126/science.1065769; PMID: 11847343
- Khan M, Rozhon W, Bigeard J, Pflieger D, Husar S, Pitzschke A, Teige M, Jonak C, Hirt H, Poppenberger B. Brassinosteroid-regulated GSK3/Shaggy-like kinases phosphorylate mitogen-activated protein (MAP) kinase kinases, which control stomata development in Arabidopsis thaliana. J Biol Chem 2013; 288:7519 - 27; http://dx.doi.org/10.1074/jbc.M112.384453; PMID: 23341468
- Tong H, Liu L, Jin Y, Du L, Yin Y, Qian Q, Zhu L, Chu C. DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 2012; 24:2562 - 77; http://dx.doi.org/10.1105/tpc.112.097394; PMID: 22685166
- Kim TW, Michniewicz M, Bergmann DC, Wang ZY. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 2012; 482:419 - 22; http://dx.doi.org/10.1038/nature10794; PMID: 22307275
- Gudesblat GE, Schneider-Pizoń J, Betti C, Mayerhofer J, Vanhoutte I, van Dongen W, Boeren S, Zhiponova M, de Vries S, Jonak C, et al. SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat Cell Biol 2012; 14:548 - 54; http://dx.doi.org/10.1038/ncb2471; PMID: 22466366
- Vert G, Walcher CL, Chory J, Nemhauser JL. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci U S A 2008; 105:9829 - 34; http://dx.doi.org/10.1073/pnas.0803996105; PMID: 18599455
- Matsui K, Umemura Y, Ohme-Takagi M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 2008; 55:954 - 67; http://dx.doi.org/10.1111/j.1365-313X.2008.03565.x; PMID: 18532977
- Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 2008; 55:940 - 53; http://dx.doi.org/10.1111/j.1365-313X.2008.03564.x; PMID: 18532978
- Ye H, Li L, Guo H, Yin Y. MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci U S A 2012; 109:20142 - 7; http://dx.doi.org/10.1073/pnas.1205232109; PMID: 23169658
- Harris JC, Hrmova M, Lopato S, Langridge P. Modulation of plant growth by HD-Zip class I and II transcription factors in response to environmental stimuli. New Phytol 2011; 190:823 - 37; http://dx.doi.org/10.1111/j.1469-8137.2011.03733.x; PMID: 21517872
- Ciarbelli AR, Ciolfi A, Salvucci S, Ruzza V, Possenti M, Carabelli M, Fruscalzo A, Sessa G, Morelli G, Ruberti I. The Arabidopsis homeodomain-leucine zipper II gene family: diversity and redundancy. Plant Mol Biol 2008; 68:465 - 78; http://dx.doi.org/10.1007/s11103-008-9383-8; PMID: 18758690
- Turchi L, Carabelli M, Ruzza V, Possenti M, Sassi M, Peñalosa A, Sessa G, Salvi S, Forte V, Morelli G, et al. Arabidopsis HD-Zip II transcription factors control apical embryo development and meristem function. Development 2013; 140:2118 - 29; http://dx.doi.org/10.1242/dev.092833; PMID: 23578926
- Carabelli M, Turchi L, Ruzza V, Morelli G, Ruberti I. Homeodomain-Leucine Zipper II family of transcription factors to the limelight: central regulators of plant development. Plant Signal Behav 2013; 8; http://dx.doi.org/10.4161/psb.25447; PMID: 23838958
- Bou-Torrent J, Salla-Martret M, Brandt R, Musielak T, Palauqui JC, Martínez-García JF, Wenkel S. ATHB4 and HAT3, two class II HD-ZIP transcription factors, control leaf development in Arabidopsis. Plant Signal Behav 2012; 7:1382 - 7; http://dx.doi.org/10.4161/psb.21824; PMID: 22918502
- Sorin C, Salla-Martret M, Bou-Torrent J, Roig-Villanova I, Martínez-García JF. ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J 2009; 59:266 - 77; http://dx.doi.org/10.1111/j.1365-313X.2009.03866.x; PMID: 19392702
- Sawa S, Ohgishi M, Goda H, Higuchi K, Shimada Y, Yoshida S, Koshiba T. The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J 2002; 32:1011 - 22; http://dx.doi.org/10.1046/j.1365-313X.2002.01488.x; PMID: 12492842
- Zhang D, Ye H, Guo H, Johnson A, Zhang M, Lin H, Yin Y. Transcription factor HAT1 is phosphorylated by BIN2 kinase and mediates brassinosteroid repressed gene expression in Arabidopsis. Plant J 2013; 77:59 - 70; http://dx.doi.org/10.1111/tpj.12368; PMID: 24164091
