Abstract
Gibberellin (GA) hormone signaling occurs through proteolytic and non-proteolytic mechanisms. GA binding to the GA receptor GID1 (GA-INSENSITIVE DWARF1) enables GID1 to bind negative regulators of GA responses called DELLA proteins. In proteolytic GA signaling, the SLEEPY1 (SLY1) F-box protein targets DELLA proteins in the GID1-GA-DELLA complex for destruction through the ubiquitin-proteasome pathway. Non-proteolytic GA signaling in sly1 mutants where GA cannot target DELLA proteins for destruction, requires GA and GID1 gene function. Based on comparison of gid1 multiple mutants to sly1 gid1 mutants, GID1a is the primary GA receptor stimulating stem elongation in proteolytic and non-proteolytic signaling, and stimulating fertility in proteolytic GA signaling. GID1b plays the primary role in fertility, and a secondary role in elongation during non-proteolytic GA signaling. The stronger role of GID1b in non-proteolytic GA signaling may result from the fact that GID1b has higher affinity for DELLA protein than GID1a and GID1c.
Keywords: :
This paper reviews evidence that the 3 Arabidopsis thaliana gibberellin (GA) hormone receptors, GID1a, GID1b, and GID1c (GIBBERELLIN-INSENSITIVE DWARF1), have partially specialized functions both in proteolytic and in the non-proteolytic GA signaling that occurs in the sly1–2 (sleepy1–2) F-box mutants (based on Ariizumi et al., and others).Citation1 The plant hormone GA stimulates seed germination, stem elongation via cell division and elongation, and the transition to flowering and fertility. GA stimulates these processes by lifting repression by the DELLA domain transcriptional regulators, through both proteolytic and non-proteolytic mechanisms.Citation1-Citation3 Proteolytic DELLA destruction requires GA hormone biosynthesis, the 3 Arabidopsis GA receptors, and the SLY1 (SLEEPY1) gene (reviewed by Hauvermale et al.).Citation4 GA stimulates the interaction of the GID1 receptors with DELLA proteins. GID1-GA-DELLA complex formation stimulates DELLA protein-protein interaction with SLY1, the F-box subunit of an SCF E3 ubiquitin ligase that polyubiquitinates DELLA proteins, thereby targeting them for destruction via the 26S proteasome. The sly1 loss-of-function mutants result in increased seed dormancy, dwarfism, and infertility associated with high level DELLA accumulation due to lack of DELLA proteolysis.Citation5 If GA signaling resulted solely from DELLA destruction, then we would expect the severity of GA-insensitive phenotypes to correlate with the level of DELLA protein accumulation. Paradoxically, sly1 mutants accumulate more DELLA protein than the GA biosynthesis mutant ga1–3, and the GA receptor gid1a gid1b gid1c triple knockout lines, but the sly1 phenotypes are not as strong as those of ga1–3 and gid1a gid1b gid1c lines. The sly1 phenotypes can be partly rescued by overexpression of each of the 3 GID1a, GID1b, and GID1c GA receptor genes on the CaMV 35S promoter, suggesting that the GID1 genes can trigger GA signaling without DELLA protein destruction.Citation1,Citation2 Non-proteolytic GA signaling requires both GA and GID1 since both the ga1–3 biosynthesis mutant and gid1 mutations exacerbate the sly1–2 phenotypes.
By comparing the previously published effects of gid1a-1, gid1b-1, and gid1c-2 mutations in the wild type Columbia (Col-0) to their effects in the sly1–2 mutant in the Col-0 background, this paper examines the relative roles of the 3 Arabidopsis GA receptor genes in proteolytic and non-proteolytic GA signaling, respectively ().Citation1,Citation6,Citation7 The role of each GID1 gene in proteolytic GA signaling can be considered by reviewing the effects of gid1a, gid1b, and gid1c single, double, and triple mutants in previous studies.Citation6-Citation8 Because DELLA proteins are not destroyed in response to GA in sly1 mutants, the sly1–2 gid1 double and triple mutants generated by Ariizumi et al.Citation1 demonstrate the relative importance of each GID1 gene in GA responses in the absence of DELLA destruction – or non-proteolytic GA signaling.
Table 1. The effects of gid1 loss-of-function and GID1-OE on GA regulated responses.
GID1a and GID1b have important roles in seed germination. In the wild-type Col-0 background, the gid1a-1 gid1b-1 gid1c-2 triple mutant was unable to germinate unless the seed coat was cut.Citation7 The gid1 single and double mutants in Col-0 were all able to germinate, suggesting that each GID1 gene can function in seed germination. However, a gid1b allele in the Nossen ecotype resulted in a decreased response to GA stimulation of seed germination, suggesting that GID1b is important for proteolytic GA signaling during seed germination.Citation8 Neither the Nossen gid1a nor Col-0 gid1c alleles were found to alter GA response during seed germination. The sly1–2 mutant is highly dormant, but acquires the ability to germinate with long (1–2 y) dry after-ripening.Citation9 The sly1–2 gid1a-1 double mutant completely failed to after-ripen in 20 mo, and the sly1–2 gid1c-2 double mutant showed reduced germination compared with sly1–2. The sly1–2 gid1b-1 double mutant seeds failed to germinate with 20 mo of after-ripening, but could not be well characterized due to limited sample size resulting from infertility. Thus, GID1a appears to be important for non-proteolytic GA signaling during sly1seed germination. Published data suggest that GID1b may be important for both proteolytic and non-protetolytic GA signaling during seed germination. But it is difficult to draw a firm conclusion of its relative importance in proteolytic and non-proteolytic GA signaling due to ecotype differences.
Phenotypic comparison of gid1 loss-of-function mutants in the sly1–2 and Col-0 wild-type backgrounds provide important insights into the functional roles of GID1a, GID1b, and GID1c in stem elongation and fertility (). In Col-0, the gid1a-1 gid1c-2 and gid1a-1 gid1c-1 double mutants are shorter than the gid1a-1 gid1b-1 line.Citation6,Citation7 In contrast, gid1a-1 gid1b-1 mutations cause a stronger decrease in sly1–2 plant height than either gid1a-1 gid1c-1 or gid1b-1 gid1c-2.Citation1 Thus, GID1a and GID1c play a stronger role in proteolytic GA signaling, whereas GID1a and GID1b together play a stronger role in non-proteolytic GA signaling during stem elongation. The gid1a-1 allele caused the strongest decrease in fertility in wild-type Col-0, whereas the gid1b-1 mutation resulted in a far stronger decrease in sly1–2 fertility than gid1a or gid1c. Thus, for fertility GID1a is more important during proteolytic, and GID1b during non-proteolytic GA signaling.
The functionality of the 3 GID1 genes can also be explored by examining how well HA:GID1 fusion constructs rescue sly1 phenotypes when overexpressed on the 35S promoter.Citation1,Citation2 While loss of GID1a function had the strongest effects on plant height and seed germination, HA:GID1b fusion protein overexpression (HA:GID1b-OE) was far more effective than HA:GID1a-OE and HA:GID1c-OE in rescuing the dwarfism and seed dormancy phenotypes of sly1–2 mutants. While loss of GID1b function caused the strongest decrease in sly1–2 fertility, HA:GID1c overexpression was most effective in rescuing the sly1–2 infertility phenotype. These results further support the idea that GID1b plays an important role in non-proteolytic GA signaling during germination and stem elongation. They also suggest that GID1c can be very effective in non-proteolytic regulation of DELLA proteins during sly1 flowering. HA:GID1b-OE also resulted a greater increase in wild-type stem elongation in the Landsberg erecta (Ler) ecotype, suggesting that GID1b can influence both proteolytic and non-proteolytic GA signaling when overexpressed.Citation2 This data demonstrates that the relative roles of the 3 GID1 genes based on loss- and gain-of-function phenotypes can be quite different from each other, suggesting that the transcriptional regulation of GID1a, GID1b, and GID1c likely contributes to determining the functional roles of GID1 genes.
The fact that HA:GID1b-OE had the strongest effect on plant height and seed germination may be explained by the fact that of the 3 GID1 proteins, GID1b has the strongest affinity for GA and DELLA protein.Citation6,Citation10,Citation11 This was previously demonstrated using yeast 2-hybrid assays, GST pulldown assays, and in vitro binding assays with purified proteins. GID1b also shows some protein-protein interaction with DELLA in the absence of GA. illustrates this point using our HA:GID1-OE constructs. More DELLA RGA protein co-immunoprecipiated with HA:GID1b than with HA:GID1a and HA:GID1c in the sly1 mutant background. Although endogenous GA is present in sly1 protein extracts, the addition of more GA increased the interaction of HA:GID1a and HA:GID1c with DELLA RGA. Given that HA:GID1b has higher affinity for DELLA, it is interesting that HA:GID1c-OE resulted in better rescue of sly1–2 fertility than did HA:GID1b-OE. This together with the fact that the sly1–2 gid1b-1 double mutant was the least fertile sly1 gid1 double mutant, suggests that GID1b protein levels must be tightly regulated during Arabidopsis flowering as having too much or too little GID1b function reduces fertility.
Figure 1. HA:GID1b co-immunoprecipitates (co-IP) more DELLA RGA than HA:GID1a and HA-GID1c. Co-IP of DELLA RGA with HA:GID1 was performed as in Ariizumi et al.Citation1 Total protein extracted from 12 d-old sly1–10 35S:HA:GID1-OE seedlings was incubated with HA agarose in the presence of 0 µM, 1 µM, or 100 µM GA3 (0.1% ethanol). Protein blot analysis was performed using anti-RGA (1:10,000), anti-HA (1:5000, Immuno Consultants Laboratory) and anti-CULLIN1 (1:10,000).Citation12,Citation13 40 µg of total protein was loaded on an SDS-PAGE gel (input). A ponceau loading control, and short (1 min) and long (10 min) exposures of the RGA blot are shown. The CUL1 blot is a negative control demonstrating the specificity of the HA:GID1 interaction with RGA.
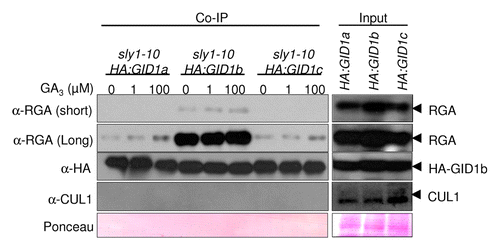
While GID1 proteins are negative regulators of DELLA protein levels during proteolytic GA signaling, GID1 genes function as positive regulators of DELLA mRNA and protein accumulation in sly1 mutants.Citation1 The gid1 mutations result in REDUCED DELLA protein and mRNA accumulation in sly1–2, suggesting that GID1 genes may normally function in positive feedback regulation of DELLA transcription. There are 5 DELLA genes in Arabidopsis. DELLA RGA (REPRESSOR OF GA1–3) regulates plant height but also participates in seed germination and fertility, whereas DELLA RGL2 (RGA-LIKE2) regulates seed germination, participates in regulating fertility, but does not regulate stem elongation. The accumulation of DELLA RGA and RGL2 proteins were examined in sly1–2 gid1 multiple mutants to examine which GID1 gene regulated each DELLA protein ().Citation1 The gid1c-2 mutation resulted in the strongest decrease in DELLA RGA accumulation in sly1 flower buds. This is consistent with the observation that HA:GID1c overexpression best rescued sly1–2 fertility (). Loss of gid1a function resulted in the strongest decrease in DELLA RGA accumulation in 4 wk-old sly1–2 plants. This is paradoxical given that sly1–2 gid1a double mutants are considerably shorter than the sly1–2 mutant that has MORE DELLA RGA repressor of plant stem elongation. Thus, in the sly1 background it is clearly not the case that more DELLA repressor correlates with shorter plants. This emphasizes the need to better understand the mechanisms underlying non-proteolytic GA signaling.
Table 2. The effects of the gid1 loss-of-function on DELLA protein accumulation in sly1–2
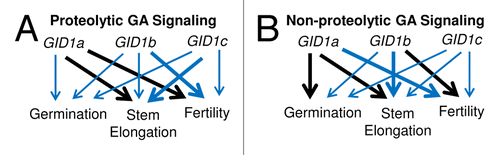
Based on comparisons of sly1 gid1 multiple mutant phenotypes to gid1 multiple mutant phenotypes, different GID1 genes predominate in non-proteolytic vs. proteolytic GA signaling (). In both cases, the GID1a gene is the main GA receptor stimulating stem elongation. However, GID1c is more important in proteolytic while GID1b is more important in non-proteolytic GA signaling during stem elongation. GID1a is the primary GA receptor stimulating fertility during proteolytic GA signaling, whereas GID1b is the primary GA receptor required during non-proteolytic GA signaling. These data suggest that GID1b protein, with its higher affinity for DELLA protein, may become more important once it is no longer possible to downregulate DELLA proteins via the ubiquitin-proteasome pathway.
Figure 2. A diagram illustrating the relative roles of the GID1 genes during (A) proteolytic, and (B) non-proteolytic GA signaling, based on gid1 loss-of-function phenotypes in the wild-type Col-0 and sly1–2 mutant backgrounds, respectively. GID1 gene function is ranked according to the severity of the gid1 loss-of-function phenotypes with heavy black arrows indicating a primary role, heavy blue arrows a secondary role, and thin blue arrows a tertiary role in each GA response.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Dr T Sun for providing the RGA antibody, Dr X Deng for providing the CULLIN1 antibody, and Dr C Schwechheimer for providing the gid1 mutant alleles in the Col-0 background. This work was supported by the National Science Foundation (award no. 0850981 to C.M.S.).
References
- Ariizumi T, Hauvermale AL, Nelson SK, Hanada A, Yamaguchi S, Steber CM. Lifting DELLA repression of Arabidopsis seed germination by non-proteolytic gibberellin signaling. Plant Physiol 2013; 162:2125 - 39; http://dx.doi.org/10.1104/pp.113.219451; PMID: 23818171
- Ariizumi T, Murase K, Sun T-P, Steber CM. Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. Plant Cell 2008; 20:2447 - 59; http://dx.doi.org/10.1105/tpc.108.058487; PMID: 18827182
- Ueguchi-Tanaka M, Hirano K, Hasegawa Y, Kitano H, Matsuoka M. Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant. Plant Cell 2008; 20:2437 - 46; http://dx.doi.org/10.1105/tpc.108.061648; PMID: 18827181
- Hauvermale AL, Ariizumi T, Steber CM. Gibberellin signaling: a theme and variations on DELLA repression. Plant Physiol 2012; 160:83 - 92; http://dx.doi.org/10.1104/pp.112.200956; PMID: 22843665
- Ariizumi T, Lawrence PK, Steber CM. The role of two F-box proteins, SLEEPY1 and SNEEZY, in Arabidopsis gibberellin signaling. Plant Physiol 2011; 155:765 - 75; http://dx.doi.org/10.1104/pp.110.166272; PMID: 21163960
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang Z-L, Powers SJ, Gong F, Phillips AL, Hedden P, Sun T-P, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 2006; 18:3399 - 414; http://dx.doi.org/10.1105/tpc.106.047415; PMID: 17194763
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EMN, Maier A, Schwechheimer C. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 2007; 19:1209 - 20; http://dx.doi.org/10.1105/tpc.107.051441; PMID: 17416730
- Iuchi S, Suzuki H, Kim Y-C, Iuchi A, Kuromori T, Ueguchi-Tanaka M, Asami T, Yamaguchi I, Matsuoka M, Kobayashi M, et al. Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. Plant J 2007; 50:958 - 66; http://dx.doi.org/10.1111/j.1365-313X.2007.03098.x; PMID: 17521411
- Ariizumi T, Steber CM. Seed germination of GA-insensitive sleepy1 mutants does not require RGL2 protein disappearance in Arabidopsis. Plant Cell 2007; 19:791 - 804; http://dx.doi.org/10.1105/tpc.106.048009; PMID: 17384169
- Nakajima M, Shimada A, Takashi Y, Kim Y-C, Park S-H, Ueguchi-Tanaka M, Suzuki H, Katoh E, Iuchi S, Kobayashi M, et al. Identification and characterization of Arabidopsis gibberellin receptors. Plant J 2006; 46:880 - 9; http://dx.doi.org/10.1111/j.1365-313X.2006.02748.x; PMID: 16709201
- Yamamoto Y, Hirai T, Yamamoto E, Kawamura M, Sato T, Kitano H, Matsuoka M, Ueguchi-Tanaka M. A rice gid1 suppressor mutant reveals that gibberellin is not always required for interaction between its receptor, GID1, and DELLA proteins. Plant Cell 2010; 22:3589 - 602; http://dx.doi.org/10.1105/tpc.110.074542; PMID: 21098733
- Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun TP. Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 2001; 13:1555 - 66; PMID: 11449051
- Wang F, Zhu D, Huang X, Li S, Gong Y, Yao Q, Fu X, Fan L-M, Deng XW. Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 2009; 21:2378 - 90; http://dx.doi.org/10.1105/tpc.108.065433; PMID: 19717618
