Abstract
With intrinsic acetyltransferase activities, CREB binding protein (CBP)/p300 proteins mediate a variety of physiological events, such as proliferation, differentiation, and apoptosis, by regulating both histones and non-histone proteins. Arabidopsis CBP-type histone acetyltransferase family proteins, HACs, have been found to influence flower by regulating the expression of Flowering Locus C. We recently reported that HAC family genes involved in the ethylene signaling pathway. Thereafter, we systematically analyzed the morphological and developmental phenotypes of all the hac mutant combinations including plant size, root, flower, leaf defects, and late-flowering. Here we reinforce the ubiquitous regulation mechanism of HAC family genes, in which HAC1 plays a dominant role with the synergistic assist of HAC5 and HAC12, whereas HAC4 slightly alleviates the influence of HAC1 and HAC5.
In mammals, CBP and p300 are very versatile proteins with intrinsic acetyltransferase activities, which participate in almost all processes of cell biology and organism development.Citation1-Citation3 On the one hand, CBP/p300 modifies histones by acetylating the lysines in their N-terminal tails, whereby relaxes the chromatin structure and make chromosomal DNA more accessible to the transcription complex.Citation4 On the other hand, structural and functional studies have shown that CBP/p300 regulates many nonhistone proteins as well, including the important transcription factors, p53 and MEF2.Citation5-Citation7 Both in vitro and in vivo data showed that CBP/p300 acetylates multiple lysine sites in the carboxyl terminus of the p53, which is indispensable for the p53-mediated transcription of p21.Citation7 Upon DNA damage, the CBP/p300 mediated acetylation of the p53 is greatly enhanced, which led to the stabilization of p53.Citation8
With a broad spectrum of CBP/p300s functions, the studies on the HAC family genes in plants just show up a tip of the iceberg. In Arabidopsis, there are 5 HAC family members, namely HAC1, HAC2, HAC4, HAC5, and HAC12. The HAT activity of HAC1 was confirmed, whereas HAC2 was proved to lack of the HAT activity. Therefore, our studies were generated in terms of HAC1, HAC4, HAC5, and HAC12. There were 2 reports demonstrated that the HAC family genes promote flower by regulating the expression of the FLC.Citation9,Citation10 Concurrently with the 2 groups, we found that hac1-involved mutants were significantly late-flowering compared with the WT plant. We measured the flowering time of all the hac mutant combinations grown under the long day condition (16h light/8h dark). We used the rosette leaf number, first flower bud appear time and first flower open time as the standards. The results were consistent for the 3 sets of data, following the sequence as WT (11.1 ± 1 RL), non-hac1-involved mutants (about 12.5 RL), hac1 (14.9 ± 1.6 RL), hac1hac4 (14.8 ± 1.6 RL), hac1hac12 (17.6 ± 2 RL), hac1hac12hac4 (18.2 ± 1.8 RL), hac1hac5 (21.9 ± 2.1 RL), hac1hac4hac5 (24 ± 2.8 RL). The results showed that all the hac1-involved mutants flower obviously later than WT, among which hac1hac5 and hac1hac4hac5 flower very late compared with WT.
We recently reported that HAC family genes influenced the ethylene signaling pathway.Citation11 The hac1-involved mutants were hypersensitive to ethylene both in the dark and in the light. Among all the hac mutant combinations, hac1hac5 mutant showed the most dramatic hypersensitivity to ethylene and hac1hac4hac5 mutant was the second behind hac1hac5. We subsequently found that the transcriptional levels of the ethylene-responsive genes were obviously higher in the hac1hac5 mutant than in the WT plant. Furthermore, our results showed that the ethylene synthesis inhibitor cannot release the classical ethylene “triple responses” of hac1hac5 and hac1hac4hac5 mutants, which excluded the possibility that hac mutations affect the ethylene sensitivity by operating the ethylene synthesis pathway. All the results led to the conclusion that HAC family genes were involved in the ethylene signaling pathway.
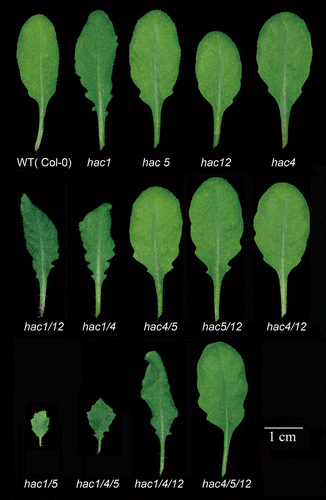
One special thought that we brought out in our previous report and that is further reinforced here, is the regulatory mechanism of the HAC family members. Knowing various functions of CBP/p300 ranging from cell growth to apoptosis, it’s not unforeseen to discover a variety of HACs’ functions. In addition to late-flowering and ethylene-hypersensitive, hac1-involved mutants displayed multiple morphological and developmental defects, such as short primary root, small plant size, short filament, and petal.Citation9-Citation11 Here we show that all the hac1-involved mutants displayed small, dark green, wrinkly, and indented rosette leaf phenotypes (). Taken together, for all the morphological and developmental phenotypes, hac1hac5 exhibited the most severe phenotypes; hac1hac4hac5 was ranked in second place, succeeded by hac1hac12 and other hac mutants. In the case of flowering time, hac1hac4hac5 flowered a bit later than hac1hac5. Considering that hac1hac5 was very weak and hac1hac4hac5 was much healthier than hac1hac5, the flowering time between these 2 mutants was not comparable. All the phenotype analysis data together suggest that HAC1, HAC12, and HAC5 function synergistically, with HAC1 being dominant and HAC5 being the closest coordinator of HAC1; HAC4 alleviates the effects of HAC1 and HAC5.
Figure 1.hac1-involved mutants display small, dark green, wrinkly, and indented leaves. The plants were grown in the long day photoperiod (16h light/8h dark) for 25 d. Among all the mutants, the hac1hac5 mutant exhibited the most severe phenotypes, followed by hac1hac4hac5, hac1hac4hac12, hac1hac12, hac1hac4, hac1, and other hac mutants.
An interesting outcome from this study was the hac1 mutation-induced dose dependent phenotypes, which was in line with the dose dependent regulatory mechanism of CBP/p300 in the embryo development.Citation11 Focusing on the protruding gynoecia and late-flowering phenotypes of the hac1 mutant, we found that the WT background transgenic plants harboring the pHAC1:HAC1 resembled the hac1 mutant; the hac1 mutant background transgenic plants harboring the 35s:HAC1 showed more severe phenotypes than hac1 mutant; the most severe phenotypes among all the HAC1 transgenic plants occurred in the WT background transgenic plants harboring the 35s:HAC1 (). These results indicated that fine tuning of the HAC1 protein levels was crucial for maintaining the homeostasis of the plant growth and development.
Table 1. Phenotype analysis of HAC1 transgenic plants
Knowing the severe morphological and developmental defects of hac mutants, it’s important to study the HAC family genes’ functions comprehensively and thoroughly. In theory, the morphological defects such as the substantial reduction of leaf and plant size would disturb the crop yield. In particular, the defective male and female gametophytes in the hac1hac5 mutant resulted in sterility. Therefore, much deeper understanding of the molecular and cellular mechanisms is required to manipulate the HAC family genes. With regard to histone targets, ChIP-on-chip could be utilized to systematically search the targets. For non-histone protein targets, yeast-2-hybrid screening based on the protein-protein interaction will be an ideal method.
| Abbreviations: | ||
| HAT | = | histone acetyltransferase |
| CBP | = | CREB-binding protein |
| HAC | = | Histone |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the Arabidopsis Biological Resources Center at the Ohio State University and Bernd Weisshaar for providing SALK and GABI T-DNA insertion lines in the Col-0 background. This work was supported by the Ministry of Science and Technology of China 863 project, Transgenic Program of China (2009ZX08009–065B), and the National Natural Science Foundation of China (31071351).
Reference
- Wang F, Marshall CB, Ikura M. Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis: structural and functional versatility in target recognition. Cell Mol Life Sci 2013; 70:3989 - 4008; http://dx.doi.org/10.1007/s00018-012-1254-4; PMID: 23307074
- Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev 2000; 14:1553 - 77; PMID: 10887150
- Giordano A, Avantaggiati ML. p300 and CBP: partners for life and death. J Cell Physiol 1999; 181:218 - 30; http://dx.doi.org/10.1002/(SICI)1097-4652(199911)181:2<218::AID-JCP4>3.0.CO;2-5; PMID: 10497301
- Görisch SM, Wachsmuth M, Tóth KF, Lichter P, Rippe K. Histone acetylation increases chromatin accessibility. J Cell Sci 2005; 118:5825 - 34; http://dx.doi.org/10.1242/jcs.02689; PMID: 16317046
- Gu W, Shi XL, Roeder RG. Synergistic activation of transcription by CBP and p53. Nature 1997; 387:819 - 23; http://dx.doi.org/10.1038/42972; PMID: 9194564
- Grossman SR. p300/CBP/p53 interaction and regulation of the p53 response. Eur J Biochem 2001; 268:2773 - 8; http://dx.doi.org/10.1046/j.1432-1327.2001.02226.x; PMID: 11358491
- Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell 2008; 133:612 - 26; http://dx.doi.org/10.1016/j.cell.2008.03.025; PMID: 18485870
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 1997; 90:595 - 606; http://dx.doi.org/10.1016/S0092-8674(00)80521-8; PMID: 9288740
- Han SK, Song JD, Noh YS, Noh B. Role of plant CBP/p300-like genes in the regulation of flowering time. Plant J 2007; 49:103 - 14; http://dx.doi.org/10.1111/j.1365-313X.2006.02939.x; PMID: 17144897
- Deng W, Liu C, Pei Y, Deng X, Niu L, Cao X. Involvement of the histone acetyltransferase AtHAC1 in the regulation of flowering time via repression of FLOWERING LOCUS C in Arabidopsis. Plant Physiol 2007; 143:1660 - 8; http://dx.doi.org/10.1104/pp.107.095521; PMID: 17416640
- Li C, Xu J, Li J, Li Q, Yang H. Involvement of Arabidopsis Histone Acetyltransferase HAC Family Genes in the Ethylene Signaling Pathway. Plant Cell Physiol 2014; 55:426 - 35; http://dx.doi.org/10.1093/pcp/pct180; PMID: 24287137
