Abstract
Vesicle transport occurs in the cytosol through a COPI, COPII and clathrin coated vesicle system for transport of lipids and proteins to different subcellular compartments. All three systems consist of several different protein components to maintain a functional transport. In chloroplasts photosynthesis takes place in thylakoids. Thylakoids contain a large amount of lipids and proteins but none of these components are produced there. Transport of lipids occurs from the envelope membrane where they are produced and through the aqueous stroma before being directed to the thylakoids. Nuclear encoded proteins use distinct pathways for entering thylakoids after import into chloroplasts. Transport of lipids through stroma requires either lipid transfer proteins, association between the envelope and the thylakoid membrane, or a vesicle transport system similar to the cytosolic one. No evidence exists for lipid transfer proteins in chloroplasts, or for a consistent association between the envelope and the thylakoid membrane. However, vesicle transport has support from e.g., biochemical and genetics data as well as transelectron microscopy data. Moreover, a recent bioinformatics study revealed COPII related proteins to be putatively chloroplast localized in Arabidopsis and thus function in vesicle transport in chloroplasts. Here we present gene expression profiles of these COPII related putatively chloroplast localized proteins using Genevestigator (https://www.genevestigator.com/gv/) with special emphasis on Rab related proteins since they represent several stages of vesicle transport e.g., uncoating, tethering and fusion.
Keywords: :
Gene Expression
The publicly available database Genevestigator (https://www.genevestigator.com/gv/) was used to identify gene expression of previously proposed chloroplast localized COPII related proteinsCitation1 at ten different developmental stages. We hypothesized that a functional chloroplast vesicle transport system containing these proteins would have gene expression linked at stages when chloroplasts are evident. However, at senescence we expect less strong correlation due to degradation of thylakoidsCitation2 and less need for vesicle transport to maintain thylakoids. We divided the predicted proteins into five groups: initiation/budding, tethering, SNARE related, Rab related, and reticulons.
We used biclustering from Genevestigator to link similar gene expression at the individual developmental stages. Four different gene expression thresholds (0%, 20%, 60% and 80%) were used to create patterns to evaluate how the groups of genes or genes within the groups related to each other during development. All genes were expressed at threshold 0% except for three genes being off at senescence, one cargo receptor and two tethering factors (COG5 and COG6) (data not shown). Twelve proteins were below the 20% threshold at senescence, three belonging to initiation/budding, five to tethering, two each for SNARE and Rab (). In addition to the tethering proteins being off at 0% four more were added when using the 20% threshold, of which three at senescence (COG3, ExoH1 and ExoH2). ExoH1 and ExoH2 appeared below the 20% threshold also at the developed rosette stage,true as well for ExoH8. ExoH8 was below the 20% threshold also at the two early stages, germinated seed and seedling, and together with ExoH1 at the bolting stage as well, whereas ExoH1 alone was under the 20% threshold at the late developmental stages of flowers and siliques and mature siliques ().
Figure 1. Biclustering of gene expression of putative chloroplast localized COPII related proteins at different developmental stages. The Arabidopsis proteins were all retrieved from a previous study where full names and more in depth information of each protein can be found.Citation1 Bioclustering data were retrieved from Genevestigator with the threshold set to 20%, black squares being above and white squares being below the threshold, respectively. Binary matrix density was 0.944.
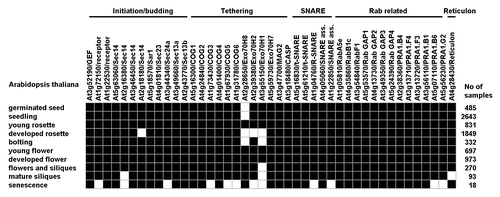
Other proteins being below the 20% threshold were two Sec14-like proteins at developed rosettes and mature siliques, respectively, and Sec24a, two SNAREs and two Rab receptors at senescence. Finally, the reticulon protein was below the 20% threshold at mature siliques (). Although several proteins were below 20% expression other proteins with same proposed function were still above, except for an R-SNARE and Sec24a for which no clear functional backup exists in the data set at senescence, the stage indicating less active phase for putative chloroplast vesicle transport components ().
Only three proteins appear above 80% expression and all at senescence, Sec13b, RabB1c and a Rab GAP2 protein (data not shown). Thus, putative chloroplast vesicle proteins are not expressed at the highest level (80%) at any stages of building thylakoids. In addition, 60% as threshold gives that most putative chloroplast vesicle proteins have gene expression between 20 to 60% since no developmental stages include necessary proteins from all groups supporting high gene expression throughout the groups (data not shown).
The group of Rab related proteins can be linked to both initial and final steps of vesicle transport.Citation3,Citation4 Thus, we scrutinized the gene expression pattern of Rab related proteins based on percent expression potential to see possible trends (). The majority have a gene expression between 33–66% () whereas three proteins were identified with a higher gene expression at the germinated seed stage including the RabA5e GTPase. It also included two Rab receptor proteins of which one of them, PRA1.F3, had high gene expression throughout all stages except for senescence where it was maximum 50% (). At the senescence stage three other proteins were observed having high expression including the two Rab GTPases, RabB1C and RabF1, and a Rab GAP2. Low gene expression (below 20%) was observed for the two mentioned above () but in addition three more had expression between 17–33% at senescence, two Rab GAPs and one Rab receptor. All Rab GAPs had 17–33% gene expression at the flowers and siliques stage, and one Rab GTPase, one Rab GAP and one Rab receptor had this at the mature siliques stage. Rab GAP4 had gene expression between 17–33% throughout all stages except for seedlings where it was slightly higher ().
Table 1. Gene expression for putative chloroplast localized Rab related proteins. The percent of gene expression for Rab related proteins at ten different individual developmental stages were highlighted giving numbers correlated with different percent interval data from Genevestigator. 1, 0–16%; 2, 17–33%; 3, 34–50%; 4, 51–66%; 5, 67–83%, 6, 84–100%
A rather homogenous gene expression exists for the groups studied (between 20–60%) at most developmental stages, indicating that necessary proteins needed to withhold a vesicle transport system could occur at stages when chloroplasts are present. The exception is senescence where most of the gene expression below 20% occurs as well as two extremes, some genes being off or above 80%. Degradation of chloroplast materials occurs at senescence proposed to include autophagy to transport materials from chloroplasts,Citation2 which could indicate a modified vesicle transport from the thylakoids. In general low gene expression at senescence for Rab related proteins in combination with other Rab related proteins having high gene expression at senescence could indicate a specific subset of Rab related proteins involved in transport from thylakoids with possible links to autophagy which also make use of vesicles.Citation2 To note, two Rab GTPases have recently been identified in chloroplasts, RabA5eCitation5 and RabF1 (unpublished observation, Aronsson’s lab), which opens up speculation for more Rab related proteins in chloroplasts. Thus, future work is needed to better understand the suggested COPII related chloroplast vesicle transport and confirm its components.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Khan NZ, Lindquist E, Aronsson H. New putative chloroplast vesicle transport components and cargo proteins revealed using a bioinformatics approach: an Arabidopsis model. PLoS One 2013; 8:e59898; http://dx.doi.org/10.1371/journal.pone.0059898; PMID: 23573218
- Ishida H, Izumi M, Wada S, Makino A.. Roles of autophagy in chloroplast recycling. Biochim Biophys Acta. 2014; 1837:512 - 21
- Nielsen E, Cheung AY, Ueda T. The regulatory RAB and ARF GTPases for vesicular trafficking. Plant Physiol 2008; 147:1516 - 26; http://dx.doi.org/10.1104/pp.108.121798; PMID: 18678743
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 2009; 10:513 - 25; http://dx.doi.org/10.1038/nrm2728; PMID: 19603039
- Karim S, Alezzawi M, Garcia-Petit C, Solymosi K, Khan NZ, Lindquist E, Dahl P, Hohmann S, Aronsson H. A novel chloroplast localized Rab GTPase protein CPRabA5e is involved in stress, development, thylakoid biogenesis and vesicle transport in Arabidopsis. Plant Mol Biol 2014; 84:675 - 92; PMID: 24337800
