Abstract
Maize KNOTTED (KN1) homeodomain (HD) protein is a well-known mobile transcription factor crucial for stem cell maintenance. Recent studies have revealed that the trihelical HD of KNOTTED1-like homeobox (KNOX) proteins is necessary and sufficient for selective cell-to-cell trafficking. Also, the efficient trafficking ability for HD is likely to be acquired during the evolution of early nonvascular land plants. Here, using the point-mutated HD of KN1 and SHOOT MERISTEMLESS (STM) in the trichome rescue system, together with molecular structure modeling, we have found the evolutionarily conserved amino acid residues, such as arginine in helix α1 and leucine in helix α3, which are essential for intercellular trafficking. Our studies provided important clues for the 3-dimensional protein structure required for cell-to-cell movement of non-cell-autonomous transcription factors.
Cell-to-cell communication is a fundamental process for multicellular organisms to coordinate developmental and environmental cues between adjacent cells.Citation1-Citation9 In higher plants, diverse cell-to-cell signaling molecules such as regulatory proteins, small RNAs, and phytohormones exchange between neighboring cells via plasmodesmata (PD).Citation10-Citation15
The proteins that traffic cell-to-cell are generally named non-cell-autonomous proteins (NCAPs). Many endogenous NCAPs, including mobile transcription factors (TFs), RNA-binding proteins and foreign viral movement proteins (MP), increase PD channel size via protein-protein interactions, and facilitate their subsequent intercellular trafficking.Citation16-Citation25 The intercellular movement of many classic non-cell-autonomous (NCA) TFs, including maize KNOTTED1 (KN1),Citation16,Citation26WUSCHEL,Citation27LEAFY,Citation28 SHORT-ROOT (SHR),Citation29,Citation30 CAPPICE, and FLOWERING LOCUS T (FT), has been observed and plays a vital role in plant development and cell fate determination.Citation31-Citation33 In addition, in Arabidopsis roots, numerous NCA TFs have been identified through genome-wide root screening systems.Citation34,Citation35 Notably, several NCAP binding regulators, including NON-CELL-AUTONOMOUS PATHWAY PROTEIN 1 (NCAPP1), the chaperonin complex, SHR INTERACTIING EMBRYONIC LETHAL (SIEL), FT-INTERACTING PROTEIN 1 (FTIP1), SCARECROW (SCR), MP binding protein 2C (MPB2C), and microtubule networks have been identified for their roles in promoting or restricting intercellular trafficking.Citation22,Citation36-Citation41
Some NCA TFs possess specific motifs or domains involved in selective movement,Citation42,Citation43 such as the intact C-terminal homeodomain (HD) of KN1 and N-terminal intercellular trafficking motif (ITM) of NCA Dof family TFs that are necessary and sufficient for intercellular transport.Citation44,Citation45 In the trichome rescue assay, the HD of KN1 or the ITM of Dof TFs confers movement capacity from mesophyll cells to epidermal cells to the cell-autonomous GLABROUS1 (GL1) protein.Citation44,Citation45 Surprisingly, the plant KN1_HD, similar to animal ENGRAILED HD protein, is able to transfer from cell to cell in mammalian cells, presumably via an unconventional secretion-internalization mechanism.Citation46 Recently, our evolutionary analysis has shown that the HD proteins of nonvascular plants acquire the capability to traffic cell-to-cell.Citation45 These signaling domains are thought to directly interact with factors involved in the NCA trafficking pathway. Intriguingly, it has been suggested that the HD of KN1 binds to the microtubule-associated protein MPB2C, which negatively regulates the trafficking ability of KN1 by tethering it to microtubules, leading to an ubiquitin-dependent 26S proteasome degradation pathway.Citation41,Citation47
Given the crucial roles played by intercellular trafficking domains and motifs in mediating protein transport and the evolution of NCA TFs, it is worthwhile to explore the relevance of evolutionarily conserved residues for interaction with the potential transport regulators in mediating cell-to-cell movement through PD. In this report, we identified 2 evolutionarily conserved amino acid residues in HD peptides that are critical for intercellular trafficking of KN1.
In a recent study, we identified homologous KNOX HD proteins based on the highly conserved HD of KN1 (KN1_HD) that is necessary and sufficient for intercellular trafficking through PD.Citation44 These HD proteins contain a conserved amino acid domain (approx. 71 aa) comprising 3 α-helices (α1, α2, and α3) and a stretch of basic lysine amino acids in front of helix α1 which has been considered as a putative nuclear localization signal (NLS) ().Citation45 Using a trichome rescue system, we previously showed that the NCA HD peptides of KNAT1, LeT6, STM, and MKNs could significantly rescue trichome development, while the cell-autonomous HD peptides of KNAT2, KNAT3, Bellringer (BLR), Gamete-specific minus 1 (GSM1), and Gamete-specific homeodomain protein 1 (HDG1) are ineffective in rescuing trichomes.Citation44,Citation45,Citation48
Figure 1. Multiple sequence alignment of homeodomain (HD) regions from KNOX homologs in maize, Arabidopsis, tomato, moss, and alga Chlamydomonas. The Accession numbers, phylogenetic trees, and intercellular trafficking activity with these HD homologs from Zea mays (KN1), Arabidopsis thaliana (KNAT1, KNAT2, KNAT3, and BLR), Lycopersicon esculentum (LeT6), Physcomitrella patens (MKN1–3, MKN2, MKN4, and MKN5), and Chlamydomonas reinhardtii (GSM1 and HDG1) are described previously.Citation44,Citation45 Residues identical or similar are shaded in black or gray, respectively, and dashes indicate gaps for optimal alignment. The elements of secondary structure such as helix α1 in violet, helix α2 in dark cyan, and helix α3 in red are denoted above the sequence. The amino acid residues highlighted by arrowheads are D29, Q30, and L53 in the corresponding helical regions. NCA, non-cell-autonomous; CA, cell-autonomous.
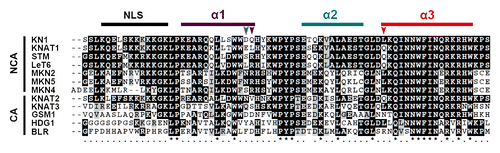
To identify the amino acid residues that are essential for intercellular trafficking, we compared the sequences of NCA HDs with those of cell-autonomous HDs. In addition, we compared sequences among NCA HDs, since KNAT1_HD, STM_HD, and LeT6_HD imparted a decreased capacity for movement compared with KN1_HD.Citation44 In general, the charged and hydrophobic residues on surface play a significant role in protein–protein interaction through ionic interaction and hydrophobic interaction, respectively. Based on the above knowledge, 2 amino acids, including R30 located at the end of helix α1 corresponding to Q30 of KN1_HD and L53 at the start of helix α3, were selected for further study (). The positively charged amino acid R30 is highly conserved in NCA HDs and differs from the corresponding residues in cell-autonomous HDs. The hydrophobic amino acid L53 in KN1 was changed to Q in other HDs, including STM and LeT6 that are relatively weak NCA TFs compared with KN1. Thus, we hypothesized that these conserved residues may play important roles for cell-to-cell trafficking of HDs.
To determine whether these residues are functionally conserved or specific for selective intercellular trafficking, the trichome rescue assay was employed to test the capacity of KN1_HD and STM_HD with equivalent mutated residues to move from mesophyll to epidermal cell layer by observing trichome development on adaxial surface of the first pair of young leaves (). In these experiments, a KN1_HD fusion to GFP-GL1 under the mesophyll-specific RbcS2b promoter (pRbcS2b::GFP-GL1-HD) was transformed into the gl1 mutant background. The GFP-GL1-HD fusion rescued trichome development in ~48% of independent T1 transgenic lines. When compared with the native KN1_HD, the mutated HD (Q30R: the Q30 at the helix α1 was replaced by the conserved residue R) significantly promoted cell-to-cell trafficking, with ~73% of transgenic seedlings showing trichome rescue. The STM_HD fusion showed 5.5% of trichome rescue, that is less efficient compared with KN1_HD fusion. When R30 was replaced by Q in the STM_HD fusion, no trichome rescue activity was detected, indicating the significance of R30 residue in cell-to-cell trafficking.
Table 1. HD mutation analysis of the capacity of intercellular trafficking in the trichome rescue system
Next, mutations at L53 in the KN1_HD fusion were tested. A mutated HD (L53A) showed a dramatically reduced percentage of trichome rescue, to about 8%. Surprisingly, a mutated KN1_HD (L53Q) fusion (L53Q: Q appears in the corresponding positions of NCA HD proteins such as STM and LeT6) did not show any trichome rescue activity. This reduction in trichome rescue activity was partly predicted, as the movement of HD peptides of STM and LeT6 is less effective than that of KN1.Citation44 In contrast, the STM_HD (Q53L) mutant increased the ratio of trichome rescue to 11.6% (50% when counted for 1st-4th leaves), compared with the native STM_HD which had a trichome rescue of 5.5% (). Accordingly, these findings indicate that the evolutionarily conserved residue L53 is important for selective intercellular trafficking of HDs. It is notable that a putative nuclear export signal (NES) sequence (residues L45-LXL53, spanning helices α2 and α3) is found in KN1 (). A similar NES plays a critical function in cell-to-cell transport of ENGRAILED2 in an animal cell system.Citation44,Citation49
To better understand the properties of the HD peptides in selective cell-to-cell trafficking, a mimic protein structure of the KN1_HD was generated. As shown in , the HD of KN1 is composed of an N-terminal stretch, 3 helices and a distinct loop conformation linking helices α1 and α2. The helices α1 and α2 tend to be located in the same horizontal plane. The position of helix α3 is far away from both of the other 2 helices, and the coil structure of the N-terminal portion from helix α1 bends over the α3 helix. Additionally, we found that the residues of helix α3 (i.e., N58, W59, I61, N62, and R66) are predicted to bind with a short TGAC core consensus sequence for KN1 proteins in a sequence-specific manner, based on the analysis through the iterative threading assembly refinement (I-TASSER) server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) ().Citation50,Citation51
Figure 2. Role of R30 at helix α1 and L53 at helix α2 in the 3-dimensional (3D) protein structure of the HD of KN1. (A) Model structure of HD of KN1 was built by the SWISS-MODEL server, using the HD of human homeobox protein TGIF2LX (2dmnA.pdb) as template structure (backbone root-mean-square deviation (rmsd): 0.40 Å ± 0.08 Å (residues 15–70)). The protein ribbon model with diverse structures is displayed using the Accelrys Discovery Studio Visualizer. Each segment of secondary structure is represented by a different color; helix α1 in violet, helix α2 in cyan, helix α3 in red, loop regions in gray, and turn regions (H31, K69, and P70) in green are marked. (B) A 3D structure of helix α3 of KN1_HD bound with DNA molecule, using various template structures through the I-TASSER server. DNA molecule is shown in dots of spheres. The amino acid residues (N58, W59, I61, N62, and R66) are marked by sticks, and labeled. (C-E) 3D protein structures of native HD compared with HD mutants and the superimposition of these proteins. The protein models of native HD (C) and double HD (Q30R and L53Q) mutant (D) were predicted using I-TASSER server. Protein structure alignment between native HD (green-colored) and double HD mutant (light blue-colored) was analyzed by PyMOL 3D-superimposition (E). Protein surface structures are highlighted on left and the corresponding cartoon structures are shown on right. Blue, positively charged amino acids; Red, negatively charged amino acids. The amino acid residues (Q30, L53, Q30R, L53Q, Q/R, and L/Q) are labeled on the surface and shown by sticks. Pockets (cavities) are marked in black arrowheads.
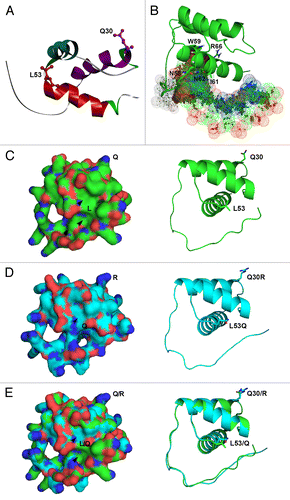
The protein structures of the KN1_HD and the related HD mutants were further studied to determine the difference of protein folding and locations of the critical residues (e.g., Q, R, and L) in stereo modes of the 3D proteins using the I-TASSER server. Visually, the protein packing of the KN1_HD clearly differed from that of KN1_HD Q30R and L53Q mutants, with the wild type being more tightly folded (). In addition, compared with the uncharged residue Q30 of the native HD, the positive charged R of the HD (Q30R) mutant protrudes from the surface of the protein (). Together with the result that the HD (Q30R) mutant facilitates intercellular trafficking (), we hypothesize that the positive charged R on the surface is advantageous for binding to potential NCA pathway proteins. Note that amino acids at the same position of some of cell-autonomous KNOX HDs are negatively charged.
More critically, the central hydrophobic residue L53 is likely linked to 2 pockets (cavities) in the native KN1_HD protein (), whereas only a shallow cavity on the upper surface is formed the in HD (L53Q) mutant (). After aligning these 2 proteins, we observed that the cavity of the HD on the upper surface is a little deeper than that of HD (L53Q) mutant (). Furthermore, we analyzed the residues around L53, and found that the dual pockets in native HD comprise 2 subsets of surrounding residues; amino acids W28, I56, F60, K69, and P70 are part of the upper cavity, and amino acids K4, L7, K54, I60, and K65 compose the deeper pocket on the lower side. The hydrophobic interactions of internally located central residues are thought to be of prime importance in determining structure, while the changing of surface residues should not affect overall structure. Thus, it is possible that the L53Q mutation in the HD could significantly affect protein folding and conformation. It has been shown that detection of pockets and cavities are especially important for their interactions with other molecules.Citation52 Since the conserved residue L53 was confirmed to be important for selective intercellular trafficking (), and formation of the dual predicted pockets (), we propose that surface residues L53 and a class of relevant hydrophobic residues within the L53-linked dual pockets are not only responsible for protein conformation, but also for functional interaction with PD-associated components and/or other non-cell-autonomous machineries.
Collectively, we conclude that the evolutionarily conserved residues R30 of helix α1 and L53 of helix α3 are important for selective intercellular trafficking of HDs in plants. The surface residues R and L, together with a class of hydrophobic residues, are capable of offering a binding interface and an ideal protein conformation for intercellular protein movement. This study suggests that the refinement of 3D protein structure modeling of NCA proteins investigated by molecular dynamics simulations and crystallization can be used to analyze a class of residues, rather than a simple sequence motif. Such classes of residues could be crucial for protein folding and mediating the interaction with potential non-cell-autonomous pathway proteins. In the future, additional mutational analysis and isolation of KN1_HD binding non-cell-autonomous pathway proteins will be necessary to figure out the KN1_HD trafficking mechanism.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were declared by authors.
Acknowledgments
We thank Y Lee (Gyeongsang National University) for rmsd calculation. This work was supported by Basic Science Research Program (NRF-2013R1A1A2007230) through the National Research Foundation of Korea funded by the Ministry of Education, the Next-Generation BioGreen 21 Program (SSAC, grant PJ009495), Rural Development Administration, Republic of Korea, and the National Science Foundation (NSF – IOS-1027003).
References
- Bloemendal S, Kück U. Cell-to-cell communication in plants, animals, and fungi: a comparative review. Naturwissenschaften 2013; 100:3 - 19; http://dx.doi.org/10.1007/s00114-012-0988-z; PMID: 23128987
- Van Norman JM, Breakfield NW, Benfey PN. Intercellular communication during plant development. Plant Cell 2011; 23:855 - 64; http://dx.doi.org/10.1105/tpc.111.082982; PMID: 21386031
- Kim I, Zambryski PC. Cell-to-cell communication via plasmodesmata during Arabidopsis embryogenesis. Curr Opin Plant Biol 2005; 8:593 - 9; http://dx.doi.org/10.1016/j.pbi.2005.09.013; PMID: 16207533
- Uchida N, Lee JS, Horst RJ, Lai HH, Kajita R, Kakimoto T, Tasaka M, Torii KU. Regulation of inflorescence architecture by intertissue layer ligand-receptor communication between endodermis and phloem. Proc Natl Acad Sci U S A 2012; 109:6337 - 42; http://dx.doi.org/10.1073/pnas.1117537109; PMID: 22474391
- Brunkard JO, Runkel AM, Zambryski PC. Plasmodesmata dynamics are coordinated by intracellular signaling pathways. Curr Opin Plant Biol 2013; 16:614 - 20; http://dx.doi.org/10.1016/j.pbi.2013.07.007; PMID: 23978390
- Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte CP, Schulze WX, Romeis T. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc Natl Acad Sci U S A 2013; 110:8744 - 9; http://dx.doi.org/10.1073/pnas.1221294110; PMID: 23650383
- Lee JY, Wang X, Cui W, Sager R, Modla S, Czymmek K, Zybaliov B, van Wijk K, Zhang C, Lu H, et al. A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell 2011; 23:3353 - 73; http://dx.doi.org/10.1105/tpc.111.087742; PMID: 21934146
- Lee JY, Lu H. Plasmodesmata: the battleground against intruders. Trends Plant Sci 2011; 16:201 - 10; http://dx.doi.org/10.1016/j.tplants.2011.01.004; PMID: 21334962
- Faulkner C, Petutschnig E, Benitez-Alfonso Y, Beck M, Robatzek S, Lipka V, Maule AJ. LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc Natl Acad Sci U S A 2013; 110:9166 - 70; http://dx.doi.org/10.1073/pnas.1203458110; PMID: 23674687
- Lucas WJ, Ham BK, Kim JY. Plasmodesmata - bridging the gap between neighboring plant cells. Trends Cell Biol 2009; 19:495 - 503; http://dx.doi.org/10.1016/j.tcb.2009.07.003; PMID: 19748270
- Stahl Y, Simon R. Gated communities: apoplastic and symplastic signals converge at plasmodesmata to control cell fates. J Exp Bot 2013; 64:5237 - 41; http://dx.doi.org/10.1093/jxb/ert245; PMID: 23975796
- Maule A, Faulkner C, Benitez-Alfonso Y. Plasmodesmata “in Communicado”. Front Plant Sci 2012; 3:30; http://dx.doi.org/10.3389/fpls.2012.00030; PMID: 22645579
- Hyun TK, Uddin MN, Rim Y, Kim JY. Cell-to-cell trafficking of RNA and RNA silencing through plasmodesmata. Protoplasma 2011; 248:101 - 16; http://dx.doi.org/10.1007/s00709-010-0225-6; PMID: 21042816
- Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol 2012; 22:125 - 32; http://dx.doi.org/10.1016/j.tcb.2011.12.001; PMID: 22260888
- Thomas CL, Bayer EM, Ritzenthaler C, Fernandez-Calvino L, Maule AJ. Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol 2008; 6:e7; http://dx.doi.org/10.1371/journal.pbio.0060007; PMID: 18215111
- Kim JY, Yuan Z, Cilia M, Khalfan-Jagani Z, Jackson D. Intercellular trafficking of a KNOTTED1 green fluorescent protein fusion in the leaf and shoot meristem of Arabidopsis. Proc Natl Acad Sci U S A 2002; 99:4103 - 8; http://dx.doi.org/10.1073/pnas.052484099; PMID: 11891300
- Wu X, Dinneny JR, Crawford KM, Rhee Y, Citovsky V, Zambryski PC, Weigel D. Modes of intercellular transcription factor movement in the Arabidopsis apex. Development 2003; 130:3735 - 45; http://dx.doi.org/10.1242/dev.00577; PMID: 12835390
- Zhou J, Wang X, Lee JY, Lee JY. Cell-to-cell movement of two interacting AT-hook factors in Arabidopsis root vascular tissue patterning. Plant Cell 2013; 25:187 - 201; http://dx.doi.org/10.1105/tpc.112.102210; PMID: 23335615
- Wu S, Gallagher KL. Mobile protein signals in plant development. Curr Opin Plant Biol 2011; 14:563 - 70; http://dx.doi.org/10.1016/j.pbi.2011.06.006; PMID: 21763178
- Kragler F, Monzer J, Xoconostle-Cázares B, Lucas WJ. Peptide antagonists of the plasmodesmal macromolecular trafficking pathway. EMBO J 2000; 19:2856 - 68; http://dx.doi.org/10.1093/emboj/19.12.2856; PMID: 10856231
- Xoconostle-Cázares B, Xiang Y, Ruiz-Medrano R, Wang HL, Monzer J, Yoo BC, McFarland KC, Franceschi VR, Lucas WJ. Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 1999; 283:94 - 8; http://dx.doi.org/10.1126/science.283.5398.94; PMID: 9872750
- Lee JY, Yoo BC, Rojas MR, Gomez-Ospina N, Staehelin LA, Lucas WJ. Selective trafficking of non-cell-autonomous proteins mediated by NtNCAPP1. Science 2003; 299:392 - 6; http://dx.doi.org/10.1126/science.1077813; PMID: 12532017
- Yoo BC, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, Lee YM, Lough TJ, Lucas WJ. A systemic small RNA signaling system in plants. Plant Cell 2004; 16:1979 - 2000; http://dx.doi.org/10.1105/tpc.104.023614; PMID: 15258266
- Ruiz-Medrano R, Xoconostle-Cazares B, Kragler F. The plasmodesmatal transport pathway for homeotic proteins, silencing signals and viruses. Curr Opin Plant Biol 2004; 7:641 - 50; http://dx.doi.org/10.1016/j.pbi.2004.09.012; PMID: 15491912
- Tilsner J, Linnik O, Louveaux M, Roberts IM, Chapman SN, Oparka KJ. Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J Cell Biol 2013; 201:981 - 95; http://dx.doi.org/10.1083/jcb.201304003; PMID: 23798728
- Kim JY, Yuan Z, Jackson D. Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development 2003; 130:4351 - 62; http://dx.doi.org/10.1242/dev.00618; PMID: 12900451
- Yadav RK, Perales M, Gruel J, Girke T, Jönsson H, Reddy GV. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev 2011; 25:2025 - 30; http://dx.doi.org/10.1101/gad.17258511; PMID: 21979915
- Sessions A, Yanofsky MF, Weigel D. Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 2000; 289:779 - 82; http://dx.doi.org/10.1126/science.289.5480.779; PMID: 10926540
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 2000; 101:555 - 67; http://dx.doi.org/10.1016/S0092-8674(00)80865-X; PMID: 10850497
- Nakajima K, Sena G, Nawy T, Benfey PN. Intercellular movement of the putative transcription factor SHR in root patterning. Nature 2001; 413:307 - 11; http://dx.doi.org/10.1038/35095061; PMID: 11565032
- Kurata T, Ishida T, Kawabata-Awai C, Noguchi M, Hattori S, Sano R, Nagasaka R, Tominaga R, Koshino-Kimura Y, Kato T, et al. Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 2005; 132:5387 - 98; http://dx.doi.org/10.1242/dev.02139; PMID: 16291794
- Jaeger KE, Wigge PA. FT protein acts as a long-range signal in Arabidopsis. Curr Biol 2007; 17:1050 - 4; http://dx.doi.org/10.1016/j.cub.2007.05.008; PMID: 17540569
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007; 316:1030 - 3; http://dx.doi.org/10.1126/science.1141752; PMID: 17446353
- Lee JY, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN. Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci U S A 2006; 103:6055 - 60; http://dx.doi.org/10.1073/pnas.0510607103; PMID: 16581911
- Rim Y, Huang L, Chu H, Han X, Cho WK, Jeon CO, Kim HJ, Hong JC, Lucas WJ, Kim JY. Analysis of Arabidopsis transcription factor families revealed extensive capacity for cell-to-cell movement as well as discrete trafficking patterns. Mol Cells 2011; 32:519 - 26; http://dx.doi.org/10.1007/s10059-011-0135-2; PMID: 22080370
- Xu XM, Wang J, Xuan Z, Goldshmidt A, Borrill PG, Hariharan N, Kim JY, Jackson D. Chaperonins facilitate KNOTTED1 cell-to-cell trafficking and stem cell function. Science 2011; 333:1141 - 4; http://dx.doi.org/10.1126/science.1205727; PMID: 21868675
- Koizumi K, Wu S, MacRae-Crerar A, Gallagher KL. An essential protein that interacts with endosomes and promotes movement of the SHORT-ROOT transcription factor. Curr Biol 2011; 21:1559 - 64; http://dx.doi.org/10.1016/j.cub.2011.08.013; PMID: 21924907
- Liu L, Liu C, Hou X, Xi W, Shen L, Tao Z, Wang Y, Yu H. FTIP1 is an essential regulator required for florigen transport. PLoS Biol 2012; 10:e1001313; http://dx.doi.org/10.1371/journal.pbio.1001313; PMID: 22529749
- Wu S, Gallagher KL. Intact microtubules are required for the intercellular movement of the SHORT-ROOT transcription factor. Plant J 2013; 74:148 - 59; http://dx.doi.org/10.1111/tpj.12112; PMID: 23294290
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 2007; 316:421 - 5; http://dx.doi.org/10.1126/science.1139531; PMID: 17446396
- Winter N, Kollwig G, Zhang S, Kragler F. MPB2C, a microtubule-associated protein, regulates non-cell-autonomy of the homeodomain protein KNOTTED1. Plant Cell 2007; 19:3001 - 18; http://dx.doi.org/10.1105/tpc.107.044354; PMID: 17965274
- Gallagher KL, Paquette AJ, Nakajima K, Benfey PN. Mechanisms regulating SHORT-ROOT intercellular movement. Curr Biol 2004; 14:1847 - 51; http://dx.doi.org/10.1016/j.cub.2004.09.081; PMID: 15498493
- Gallagher KL, Benfey PN. Both the conserved GRAS domain and nuclear localization are required for SHORT-ROOT movement. Plant J 2009; 57:785 - 97; http://dx.doi.org/10.1111/j.1365-313X.2008.03735.x; PMID: 19000160
- Kim JY, Rim Y, Wang J, Jackson D. A novel cell-to-cell trafficking assay indicates that the KNOX homeodomain is necessary and sufficient for intercellular protein and mRNA trafficking. Genes Dev 2005; 19:788 - 93; http://dx.doi.org/10.1101/gad.332805; PMID: 15805469
- Chen H, Ahmad M, Rim Y, Lucas WJ, Kim JY. Evolutionary and molecular analysis of Dof transcription factors identified a conserved motif for intercellular protein trafficking. New Phytol 2013; 198:1250 - 60; http://dx.doi.org/10.1111/nph.12223; PMID: 23506539
- Tassetto M, Maizel A, Osorio J, Joliot A. Plant and animal homeodomains use convergent mechanisms for intercellular transfer. EMBO Rep 2005; 6:885 - 90; http://dx.doi.org/10.1038/sj.embor.7400487; PMID: 16113652
- Curin M, Ojangu EL, Trutnyeva K, Ilau B, Truve E, Waigmann E. MPB2C, a microtubule-associated plant factor, is required for microtubular accumulation of tobacco mosaic virus movement protein in plants. Plant Physiol 2007; 143:801 - 11; http://dx.doi.org/10.1104/pp.106.091488; PMID: 17189338
- Lee JH, Lin H, Joo S, Goodenough U. Early sexual origins of homeoprotein heterodimerization and evolution of the plant KNOX/BELL family. Cell 2008; 133:829 - 40; http://dx.doi.org/10.1016/j.cell.2008.04.028; PMID: 18510927
- Maizel A, Bensaude O, Prochiantz A, Joliot A. A short region of its homeodomain is necessary for engrailed nuclear export and secretion. Development 1999; 126:3183 - 90; PMID: 10375508
- Bolduc N, Hake S, Jackson D. Dual functions of the KNOTTED1 homeodomain: sequence-specific DNA binding and regulation of cell-to-cell transport. Sci Signal 2008; 1:pe28; http://dx.doi.org/10.1126/scisignal.123pe28; PMID: 18544748
- Smith HMS, Boschke I, Hake S. Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc Natl Acad Sci U S A 2002; 99:9579 - 84; http://www.pnas.prg/cgl/doi/10.1073/pnas.092271599 http://dx.doi.org/10.1073/pnas.092271599; PMID: 12093897
- Yu J, Zhou Y, Tanaka I, Yao M. Roll: a new algorithm for the detection of protein pockets and cavities with a rolling probe sphere. Bioinformatics 2010; 26:46 - 52; http://dx.doi.org/10.1093/bioinformatics/btp599; PMID: 19846440
