Abstract
The microRNA319 family (miR319) is one of the most conserved and ancient microRNA (miRNA) families in plants. Transgenic creeping bentgrass (Agrostis stolonifera) overexpressing a rice miR319, Osa-miR319a, exhibited enhanced salt and drought tolerance. A comprehensive hypothetical model about the role of miR319 in creeping bentgrass response to salinity and drought stress was proposed. Salinity and drought stress induces elevated expression of miR319, resulting in downregulation of at least 4 putative target genes of miR319 (AsPCF5, AsPCF6, AsPCF8, and AsTCP14) as well as a homolog of the rice NAC domain gene AsNAC60, and therefore positively contributing to plant abiotic stress response. Hormones might also regulate miR319 and its targets, and the expression level of the miR319 targets might be a balance of miR319-mediated target cleavage and hormone regulation of the targets. Furthermore, HKT gene families involved in salt exclusion mechanisms as well as mechanisms controlling the timing of gene expression network are also hypothesized to play an important role in this pathway.
The microRNA319 (miR319) family, one of the most conserved and ancient microRNA (miRNA) family, has previously been found to be responsive to multiple stresses including salinity, drought, cold, and aluminum stress based on high-throughput sequencing and microarray profiling data.Citation1-Citation6 In recent years, an increasing number of studies on how miR319 is involved in plant stress response were conducted to define the function of miR319. For example, a hypothetical model of how miR319 regulates plant cold stress response in sugarcane was proposed.Citation5 In this model, miR319 was upregulated by both cold stress and phytohormone abscisic acid (ABA) treatment, which caused repression of its targets leading to inhibition of plant development. It was also hypothesized that ABA might induce miR319 targets, and that the final expression level of miR319 targets was a balance of miR319-mediated cleavage and ABA induction of the targets.Citation5 Interestingly, a recent study in riceCitation7 demonstrated that overexpression of miR319 led to enhanced plant cold tolerance (4 °C) after chilling acclimation (12 °C), and that the expression level of the five target genes of miR319 (OsPCF5, OsPCF6, OsPCF7, OsPCF8, and OsTCP21) all decreased in transgenic plants. It was further demonstrated that downregulation of either one of the 2 target genes, OsPCF5 or OsPCF8 via RNA interference (RNAi) also resulted in enhanced cold tolerance in transgenic plants.Citation7 We have investigated the role of miR319 in plant abiotic stress resistance using transgenic creeping bentgrass (Agrostis stolonifera) and found that miR319 regulates plant response to drought and salinity stress via downregulation of its target genes.Citation8
Cold stress causes mechanical damages, while salinity and drought stresses mainly impose osmotic and ionic pressure on plant cells.Citation9 In nature, plants usually have to combat with a combination of multiple stresses instead of a single one, which requires crosstalk of various signals and coordination of different stress-responsive pathways.Citation9 Based on the above-mentioned potential involvement of miR319 in diverse stressors, it is plausible to postulate that miR319 could be one of the most important master regulators involved in different stress-responsive pathways. Here we propose a hypothetical model of the molecular mechanisms of miR319-mediated plant salinity and drought stress tolerance in creeping bentgrass ().
Figure 1. Hypothetical model of molecular mechanisms of miR319-mediated plant salinity and drought stress tolerance in creeping bentgrass. The accumulation of mature miR319 was elevated upon salt and drought stress, which causes downregulation of at least 4 putative target genes of miR319 (AsPCF5, AsPCF6, AsPCF8, and AsTCP14) as well as a homolog of the rice NAC domain gene AsNAC60, and therefore positively contributing to plant abiotic stress response. Hormones might also regulate miR319 and its targets, and the expression level of the miR319 targets might be a balance of miR319-mediated target cleavage and hormone regulation of the targets. Furthermore, HKT gene families involved in salt exclusion mechanisms as well as mechanisms controlling the timing of gene expression network are also hypothesized to play an important role in this pathway.
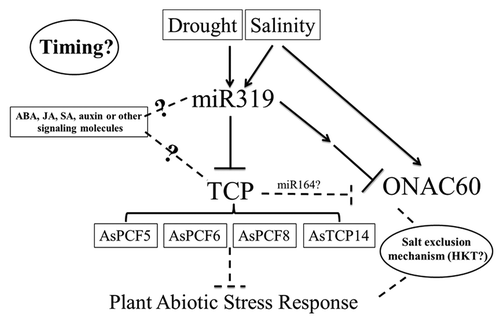
In creeping bentgrass, the accumulation of mature miR319 was elevated upon salt and drought stress, which caused the downregulation of at least 4 putative target genes of miR319 (AsPCF5, AsPCF6, AsPCF8, and AsTCP14) ().Citation8 These target genes were hypothesized to serve as negative regulators of plant abiotic stress tolerance and plant development. We also examined what other stress-related genes are involved in miR319-mediated stress tolerance pathway. A previous study analyzing stress-responsive genes in rice has revealed that a member of NAC family (NAM from petunia, ATAF and CUC from Arabidopsis), named ONAC60 (Os12 g41680) was significantly upregulated by salt stress.Citation10 ONAC60 has been known to be the target gene of miR164 in rice,Citation11 and TCP genes have been shown to regulate miR164 in Arabidopsis.Citation12 Theoretically, if this mechanism is conserved across plant species, the expression level of AsNAC60, our newly identified ONAC60 homolog in creeping bentgrass, would be expected to increase in transgenic plants overexpressing miR319 via miR319 repression of TCP-mediated miR164 regulation pathway. Surprisingly, the expression level of AsNAC60 in transgenic creeping bentgrass plants was decreased (),Citation8 suggesting that the regulation of NAC60 expression is species-dependent. It is also possible that AsNAC60 might be downregulated by other more powerful regulators. The functions of ONAC60 have not been fully characterized in model plant species rice, and remain to be investigated.
TCPs have been demonstrated to regulate jasmonic acid (JA) synthesis and leaf senescence in Arabidopsis,Citation13 while miR319 was found to be responsive to ABA treatment in sugarcane,Citation5 to positively regulate auxin (AUX) signals in rice and Arabidopsis,Citation14-Citation16 and to be involved in coordinating the antagonistic functions of AUX and cytokinin (CK) pathways and gibberellic acid (GA) and ABA pathways.Citation14,Citation17,Citation18 Furthermore, analysis of upstream promoter sequence of miR319 in Arabidopsis has revealed the existence of stress-related elements including MBS (drought), HSE (heat stress), AREs (hypoxic, low temperature, and dehydration stress), TC-rich repeats (defense and stress), TGA element (auxin), AuxRR core (auxin), and TCA element (SA).Citation2 The hormones JA, salicylic acid (SA), AUX, and ABA have all been postulated or shown to play an important role directly or indirectly in plant response to abiotic stress.Citation19 Thus, it is intriguing to assume that in creeping bentgrass, miR319 also played an important role in phytohormone crosstalk and abiotic stress signaling transduction. Additionally, the expression of miR319 and its targets might be subjected to regulations of several phytohormones or other regulators. The ultimate expression levels of these genes reflect the homeostasis equilibrium of multiple factors ().
Our results in creeping bentgrass suggested that transgenic plants overexpressing miR319 might adopt salt exclusion mechanisms to improve its salinity tolerance.Citation8 Coincidently, in transgenic rice plants overexpressing miR319, the Na+ transporter gene OsHKT2 was upregulated (Yang C, Li D, Zhu L, personal communication). It would be interesting to investigate whether any expression changes in HKT gene families take place in transgenic creeping bentgrass overexpressing miR319. This would allow better understanding of the miR319-mediated salt exclusion mechanism ().
In rice, the more rapid response of Osa-miR319 genes and their targets to low temperature was associated with improved plant cold tolerance.Citation7 In contrast, the expression change of sugarcane miR319 was delayed in cold tolerant cultivars.Citation5 These data indicated that besides expression level changes in miR319 and their targets, timing was also critical for coordinated interactions of multiple regulators determining plant stress response. Further investigation in this direction would provide information to substantiate the current hypothetical model of miR319-mediated plant stress response pathway ().
| Abbreviations: | ||
| miRNA | = | microRNA |
| miR319 | = | microRNA319 |
| ABA | = | abscisic acid |
| AUX | = | auxin |
| CK | = | cytokinin |
| ET | = | ethylene |
| GA | = | gibberellic acid |
| JA | = | jasmonic acid |
| SA | = | salicylic acid |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by Biotechnology Risk Assessment Grant Program competitive grant no. 2007–33522–18489 and no. 2010–33522–21656 from the USDA National Institute of Food and Agriculture as well as the USDA grant CSREES SC-1700315 and SC-1700450. Technical Contribution No. 6219 of the Clemson University Experiment Station.
References
- Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004; 16:2001 - 19; http://dx.doi.org/10.1105/tpc.104.022830; PMID: 15258262
- Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana.. RNA 2008; 14:836 - 43; http://dx.doi.org/10.1261/rna.895308; PMID: 18356539
- Lv DK, Bai X, Li Y, Ding XD, Ge Y, Cai H, Ji W, Wu N, Zhu YM. Profiling of cold-stress-responsive miRNAs in rice by microarrays. Gene 2010; 459:39 - 47; http://dx.doi.org/10.1016/j.gene.2010.03.011; PMID: 20350593
- Zhou L, Liu Y, Liu Z, Kong D, Duan M, Luo L. Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa.. J Exp Bot 2010; 61:4157 - 68; http://dx.doi.org/10.1093/jxb/erq237; PMID: 20729483
- Thiebaut F, Rojas CA, Almeida KL, Grativol C, Domiciano GC, Lamb CRC, Engler JdeA, Hemerly AS, Ferreira PCG. Regulation of miR319 during cold stress in sugarcane. Plant Cell Environ 2012; 35:502 - 12; http://dx.doi.org/10.1111/j.1365-3040.2011.02430.x; PMID: 22017483
- Chen L, Wang T, Zhao M, Tian Q, Zhang WH. Identification of aluminum-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. Planta 2012; 235:375 - 86; http://dx.doi.org/10.1007/s00425-011-1514-9; PMID: 21909758
- Yang C, Li D, Mao D, Liu X, Ji C, Li X, Zhao X, Cheng Z, Chen C, Zhu L. Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ 2013; 36:2207 - 18; http://dx.doi.org/10.1111/pce.12130; PMID: 23651319
- Zhou M, Li D, Li Z, Hu Q, Yang C, Zhu L, Luo H. Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol 2013; 161:1375 - 91; http://dx.doi.org/10.1104/pp.112.208702; PMID: 23292790
- Mahajan S, Tuteja N. Cold, salinity and drought stresses: an overview. Arch Biochem Biophys 2005; 444:139 - 58; http://dx.doi.org/10.1016/j.abb.2005.10.018; PMID: 16309626
- Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010; 465:30 - 44; http://dx.doi.org/10.1016/j.gene.2010.06.008; PMID: 20600702
- Wu L, Zhang Q, Zhou H, Ni F, Wu X, Qi Y. Rice MicroRNA effector complexes and targets. Plant Cell 2009; 21:3421 - 35; http://dx.doi.org/10.1105/tpc.109.070938; PMID: 19903869
- Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 2010; 22:3574 - 88; http://dx.doi.org/10.1105/tpc.110.075598; PMID: 21119060
- Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 2008; 6:e230; http://dx.doi.org/10.1371/journal.pbio.0060230; PMID: 18816164
- Curaba J, Singh MB, Bhalla PL. miRNAs in the crosstalk between phytohormone signalling pathways. J Exp Bot 2014; 65:1425 - 38; http://dx.doi.org/10.1093/jxb/eru002; PMID: 24523503
- Kant S, Bi YM, Zhu T, Rothstein SJ. SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol 2009; 151:691 - 701; http://dx.doi.org/10.1104/pp.109.143875; PMID: 19700562
- Tian Q, Uhlir NJ, Reed JW. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 2002; 14:301 - 19; http://dx.doi.org/10.1105/tpc.010283; PMID: 11884676
- Liu Q, Zhang YC, Wang CY, Luo YC, Huang QJ, Chen SY, Zhou H, Qu LH, Chen YQ. Expression analysis of phytohormone-regulated microRNAs in rice, implying their regulation roles in plant hormone signaling. FEBS Lett 2009; 583:723 - 8; http://dx.doi.org/10.1016/j.febslet.2009.01.020; PMID: 19167382
- Srivastava S, Srivastava AK, Suprasanna P, D’Souza SF. Identification and profiling of arsenic stress-induced microRNAs in Brassica juncea.. J Exp Bot 2013; 64:303 - 15; http://dx.doi.org/10.1093/jxb/ers333; PMID: 23162117
- Peleg Z, Blumwald E. Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol 2011; 14:290 - 5; http://dx.doi.org/10.1016/j.pbi.2011.02.001; PMID: 21377404
