Abstract
Plant vacuoles are essential and dynamic organelles, and mechanisms of vacuole biogenesis and fusion are not well characterized. We recently demonstrated that Wortmannin, an inhibitor of Phosphatidylinositol 3-Kinase (PI3K), induces the fusion of plant vacuoles both in roots of itt3/vti11 mutant alleles and in guard cells of wild type Arabidopsis and Fava bean. Here we used Fluorescence Recovery After Photobleaching (FRAP) to demonstrate that the vacuoles in itt3/vti11 are independent organelles. Furthermore, we used fluorescent protein reporters that bind specifically to Phosphatidylinositol 3-Phosphate (PtdIns(3)P) or PtdIns(4)P to show that Wortmannin treatments that induce the fusion of vti11 vacuoles result in the loss of PtdIns(3)P from cellular membranes. These results provided supporting evidence for a critical role of PtdIns(3)P in vacuole fusion in roots and guard cells.
The vacuole is an essential organelle that is critical for cellular homeostasis, establishment of turgor pressure and storage.Citation1-5 The molecular mechanisms for plant vacuole biogenesis and fusion are not fully understood. A pathway for pro-vacuole formation from ER membranes was recently shown in meristematic root cells in Arabidopsis.Citation6 Lytic vacuoles may also form by maturation and fusion of protein storage vacuoles as it was visualized in developing root tips during tobacco germination.Citation7 We recently demonstrated that the VTI11 SNARE protein is critical for the maintenance or biogenesis of the large central vacuole in plant cells.Citation8 VTI11 is a vacuolar SNARE protein involved in membrane fusion that was shown to regulate gravitropism and protein trafficking to the vacuole.Citation9-11 vti11 mutant alleles such as impaired tonoplast trafficking3 (itt3) display defects in vacuole fusion both during root and hypocotyl development, and during the formation of a large vacuole in guard cells.Citation8 It is likely that the vacuolar SNARE complex containing VTI11,Citation9 which localizes to the pre-vacuolar compartment (PVC) and vacuole,Citation9,12 regulates vacuole fusion events during the formation of the large central vacuole.
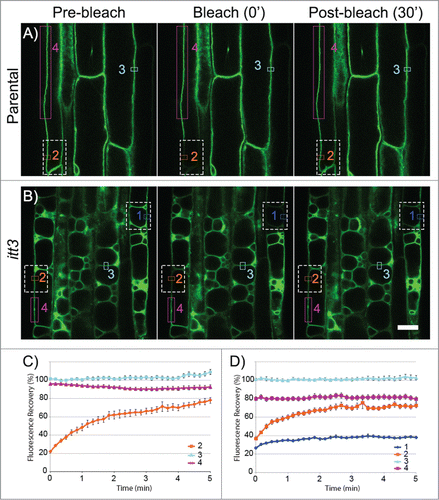
Recent surface renderings of stained vacuoles in Arabidopsis meristematic cells showed tubular interconnected vacuolar compartments that form a single organelle.Citation6 Therefore, we questioned whether the multiple vacuole phenotype in vti11/itt3 mutantsCitation8 resulted from independent or interconnected vacuoles. We used Fluorescent Recovery after Photobleaching (FRAP) to differentiate between these two possibilities. Bleaching a region of the vacuole in the parental control resulted in almost complete fluorescence recovery (, orange). Similarly, bleaching only a fraction of one of the itt3 vacuoles results in fluorescence recovery due to the movement of GFP-TIP2;1 from non-bleached regions in that vacuole (, orange). In contrast, vacuoles that are completely bleached recover poorly to the pre-bleached levels even after 5 min (, dark blue). Vacuoles that were not bleached or regions outside the bleach area of partially bleached vacuoles were included as controls (). To quantify these results, 36 FRAP experiments with root epidermal cells were analyzed and fluorescence recovery was quantified based on relative fluorescence intensity (). Partially bleached vacuoles in itt3, and the parental control recovered to almost 80% of the original fluorescence intensity. In contrast, membranes from completely bleached vacuoles in itt3 did not recover. These results indicate that vti11/itt3 vacuoles are independent compartments and not connected to adjacent vacuoles.
Figure 1. itt3 vacuoles are independent organelles. (A–B) Root epidermal cells from GFP-TIP2;1 (parental control, A) or itt3 (B) were used for FRAP using a Zeiss710 confocal microscope and images were captured every 10 s. Images before (pre-bleach), immediately after (bleach 0’) and 6 min after (post-bleach 30’) bleaching are shown for one experiment. Bleached areas are shown with white (dashed) rectangles. ROIs that were used to measure fluorescence recovery are shown with colored rectangles. ROI fluorescence was quantified for complete vacuoles included in the bleach area (1, dark blue), vacuoles partially included in the bleach area (2, orange), non-bleached controls (3, light blue), and an area adjacent to the bleach area in partially bleached vacuoles (4, magenta). To measure the fluorescence recovery of vacuoles that were completely bleached, only the membrane adjacent to the cell wall was selected for quantification (1, dark blue). Bleaching was accomplished with an argon laser in the Zeiss LSM 710 microscope with excitation wavelength of 488 nm. The laser was used at 100% power and the pixel dwell time was 100.85 μsec. Scale bar: 20 μm. (C–D) Quantification of fluorescence recovery over time for GFP-TIP2;1 (parental control, C) or itt3 (D). Using 36 sets of FRAP experiments for each genotype as shown in A–C, the percent fluorescence recovery was calculated for each region of interest. The numbers and colors in the graphs correspond to those in A–B. N: 7 seedlings. Error bars indicate standard error.
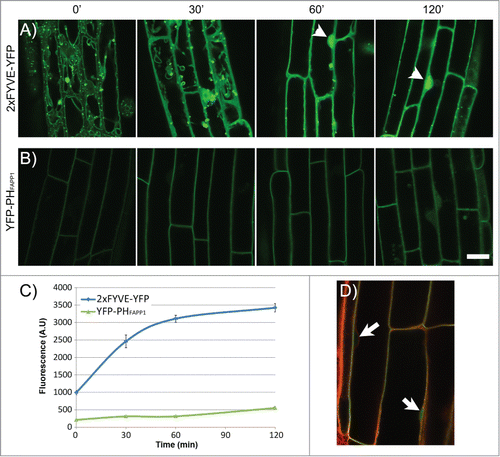
Vacuole fusion in plants is also regulated by phosphoinositides.Citation8 Using a pharmacological approach, Zheng et al. showed that inhibition of PtdIns(3)P synthesis by either Wortmannin or LY294002 is sufficient to induce vacuole fusion of itt3 vacuoles in Arabidopsis roots and fragmented vacuoles of closed guard cells in Fava bean.Citation8 Given that most mature vegetative cells maintain a single large vacuole, an effect of Phosphatidylinositol 3-Kinase (PI3K) inhibitors on vacuole fusion could not previously be observed, and it was the prevalent view that Wm induced fusion only of PVCs.Citation13 The effect of Wm on vacuole fusion indicated a critical role for phosphoinositides in regulating very dynamic changes in vacuole morphology.Citation8 Furthermore, loss of function of SUPPRESSOR OF ACTIN 2–5 (SAC2–5), results in abnormal phosphoinositide levels and changes in vacuole morphology.Citation14 A challenge for these analyses is that the loss of PI3K function is gametophytic lethal.Citation15 In addition, manipulating phosphoinositide levels by either chemical inhibitors or genetically may result in changes of multiple forms of these lipids due to lack of specificity or inter-conversion between different species.Citation16-18 In the case of Zheng et al.,Citation8 we wanted to investigate the effect of Wm on PtdIns(3)P accumulation in cells from the root elongation zone. In order to test this, we used two biosensors that specifically bind to and report the levels of two phosphoinositides, YFP-2xFYVE to visualize PtdIns(3)PCitation19 and YFP-PHFAPP1 to visualize PtdIns(4)P.Citation20 The effect of Wm on the localization of YFP-2xFYVE has been tested in tobacco cellsCitation19 and Arabidopsis root tips;Citation21 however, these data were not available for mature root cells where the central vacuole is fully formed, and where vacuole fusion events can be visualized in the itt3 mutant.Citation8 YFP-PHFAPP1 has been shown to localize to Golgi and plasma membrane,Citation20,22 and Wm did not affect the localization of this marker in tobacco BY-2 cells.Citation20 Similar to Zheng et al.,Citation8 we exposed the two marker lines to 33 μM Wm and imaged mature root cells by confocal scanning laser microscopy. The YFP-2xFYVE marker localizes to the tonoplast as well as punctate compartments, most likely pre-vacuolar compartments as previously reported ().Citation19 After 30 min of Wm treatment, the YFP-2xFYVE fluorescence shifted to the cytosol as shown by the more diffused signal between the vacuole and the cell periphery and the bright signal inside the nucleus. A defined tonoplast membrane signal is difficult to discern at 60 and 120 min (). The loss of YFP-2xFYVE from endomembranes indicates a reduction in PtdIns(3)P levels that correlates with the inhibition of PI3-Kinase. The accumulation of the fluorescent marker in the cytosol is similar to the changes in YFP-2xFYVE fluorescence in tobacco BY-2 cells treated with Wm.Citation19 In contrast, the PtdIns(4)P sensor, which is abundantly localized to the plasma membrane in the control, does not change significantly at 30 min of Wm treatment (). To indirectly determine the accumulation of the two sensors in the soluble fraction, e.g., no longer associated with a membrane, we quantified the fluorescence intensity of each marker inside the nucleus. Labeling with Lysotracker Red was used to identify the position of nucleus (). As shown in , the nuclear signal of 2xFYVE-YFP increases significantly at 30 min of Wm treatment while that of YFP-PHFAPP1 increases only at 2 h. Results from these experiments indicate that Wm treatment of Arabidopsis roots results in a significant decrease of PtdIns(3)P in tonoplast membranes within 30 min, but not in major changes in PtdIns(4)P at this time point. This timing correlates well with the timing of fusion events in itt3 roots,Citation8 and provides further supportive evidence for a specific role of PtdIns(3)P on vacuole fusion in plants. Our hypothesis is that the multiple independent vacuoles in itt3/vti11 result from delayed homotypic vacuole fusion during early stages of seedling germination that require VTI11 SNARE function and the regulation of PtdIns(3)P in the tonoplast.
Figure 2. Effect of Wortmannin on the localization of YFP-2xFYVE and YFP-PHFAPP1 in the Arabidopsis root. (A–B) Time-lapse imaging of plant roots expressing YFP-2xFYVE (A) or YFP-PHFAPP1 (B) by confocal microscopy after Wm treatment. Seedlings were incubated for 0–120 min with 33 μM Wm. Acquisition settings were kept constant throughout the experiment in order to compare protein abundance between different time points. Bright nuclear signal is indicated with arrowheads. All images were captured on a Zeiss LSM710 confocal microscope. Scale bar: 20 μm. (C–D) Quantification of fluorescence signal in the nucleus in the 2xFYVE-YFP and YFP-PHFAPP1 during Wm treatment. Seedlings were treated as in (A) and stained with Lysotracker Red for 30 min before imaging. Nuclear signal for each marker was quantified using Zen software (Zeiss). N: 20 cells, 3 seedlings per data point. Bars represent standard error. (D) Lysotracker Red staining of the YFP-PHFAPP1 line to locate the nuclei (arrows).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank W. Boss for critical reading of the manuscript.
Funding
This work was supported by National Science Foundation grant MCB-1244354 and NASA grant NNX13AM49G to M.R.P.
References
- Xiang L, Etxeberria E, Van den Ende W. Vacuolar protein sorting mechanisms in plants. FEBS J 2013; 280:979-93; PMID:23241209; http://dx.doi.org/10.1111/febs.12092
- Park M, Jurgens G. Membrane traffic and fusion at post-Golgi compartments. Front Plant Sci 2012; PMID:22645561
- Fujimoto M, Ueda T. Conserved and plant-unique mechanisms regulating plant post-Golgi traffic. Front Plant Sci 2012; 3:197; PMID:22973281; http://dx.doi.org/10.3389/fpls.2012.00197
- Bassham DC, Brandizzi F, Otegui MS, Sanderfoot AA. The secretory system of Arabidopsis. The Arabidopsis Book, 2008:1-29.
- Pedrazzini E, Komarova NY, Rentsch D, Vitale A. Traffic routes and signals for the tonoplast. Traffic 2013; 14:622-8; PMID:23356396; http://dx.doi.org/10.1111/tra.12051
- Viotti C, Krüger F, Krebs M, Neubert C, Fink F, Lupanga U, Scheuring D, Boutté Y, Frescatada-Rosa M, Wolfenstetter S, et al. The endoplasmic reticulum is the main membrane source for biogenesis of the lytic vacuole in Arabidopsis. Plant Cell 2013; 25:3434-49; PMID:24014545; http://dx.doi.org/10.1105/tpc.113.114827
- Zheng H, Staehelin LA. Protein storage vacuoles are transformed into lytic vacuoles in root meristematic cells of germinating seedlings by multiple, cell type-specific mechanisms. Plant Physiol 2011; 155:2023-35; PMID:21278307; http://dx.doi.org/10.1104/pp.110.170159
- Zheng J, Han SW, Rodriguez-Welsh MF, Rojas-Pierce M. Homotypic vacuole fusion requires VTI11 and is regulated by phosphoinositides. Mol Plant 2014; 7:1026-40; PMID:24569132; http://dx.doi.org/10.1093/mp/ssu019
- Ebine K, Okatani Y, Uemura T, Goh T, Shoda K, Niihama M, Morita MT, Spitzer C, Otegui MS, Nakano A, et al. A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell 2008; 20:3006-21; PMID:18984676; http://dx.doi.org/10.1105/tpc.107.057711
- Sanderfoot AA, Kovaleva V, Bassham DC, Raikhel NV. Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol Biol Cell 2001; 12:3733-43; PMID:11739776; http://dx.doi.org/10.1091/mbc.12.12.3733
- Yano D, Sato M, Saito C, Sato MH, Morita MT, Tasaka M. A SNARE complex containing SGR3/AtVAM3 and ZIG/VTI11 in gravity-sensing cells is important for Arabidopsis shoot gravitropism. Proc Natl Acad Sci U S A 2003; 100:8589-94; PMID:12815100; http://dx.doi.org/10.1073/pnas.1430749100
- Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH. Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct Funct 2004; 29:49-65; PMID:15342965; http://dx.doi.org/10.1247/csf.29.49
- Wang J, Cai Y, Miao Y, Lam SK, Jiang L. Wortmannin induces homotypic fusion of plant prevacuolar compartments. J Exp Bot 2009; 60:3075-83; PMID:19436047; http://dx.doi.org/10.1093/jxb/erp136
- Nováková P, Hirsch S, Feraru E, Tejos R, van Wijk R, Viaene T, Heilmann M, Lerche J, De Rycke R, Feraru MI, et al. SAC phosphoinositide phosphatases at the tonoplast mediate vacuolar function in Arabidopsis. Proc Natl Acad Sci U S A 2014; 111:2818-23; PMID:24550313; http://dx.doi.org/10.1073/pnas.1324264111
- Lee Y, Kim ES, Choi Y, Hwang I, Staiger CJ, Chung YY, Lee Y. The Arabidopsis phosphatidylinositol 3-kinase is important for pollen development. Plant Physiol 2008; 147:1886-97; PMID:18515640; http://dx.doi.org/10.1104/pp.108.121590
- Munnik T, Nielsen E. Green light for polyphosphoinositide signals in plants. Curr Opin Plant Biol 2011; 14:489-97; PMID:21775194; http://dx.doi.org/10.1016/j.pbi.2011.06.007
- Thole JM, Nielsen E. Phosphoinositides in plants: novel functions in membrane trafficking. Curr Opin Plant Biol 2008; 11:620-31; PMID:19028349; http://dx.doi.org/10.1016/j.pbi.2008.10.010
- Balla T, Szentpetery Z, Kim YJ. Phosphoinositide signaling: new tools and insights. Physiology (Bethesda) 2009; 24:231-44; PMID:19675354; http://dx.doi.org/10.1152/physiol.00014.2009
- Vermeer JE, van Leeuwen W, Tobeña-Santamaria R, Laxalt AM, Jones DR, Divecha N, Gadella TW Jr., Munnik T. Visualization of PtdIns3P dynamics in living plant cells. Plant J 2006; 47:687-700; PMID:16856980; http://dx.doi.org/10.1111/j.1365-313X.2006.02830.x
- Vermeer JE, Thole JM, Goedhart J, Nielsen E, Munnik T, Gadella TW Jr. Imaging phosphatidylinositol 4-phosphate dynamics in living plant cells. Plant J 2009; 57:356-72; PMID:18785997; http://dx.doi.org/10.1111/j.1365-313X.2008.03679.x
- Takáč T, Pechan T, Samajová O, Ovečka M, Richter H, Eck C, Niehaus K, Samaj J. Wortmannin treatment induces changes in Arabidopsis root proteome and post-Golgi compartments. J Proteome Res 2012; 11:3127-42; PMID:22524784; http://dx.doi.org/10.1021/pr201111n
- Thole JM, Vermeer JE, Zhang Y, Gadella TW Jr., Nielsen E. Root hair defective4 encodes a phosphatidylinositol-4-phosphate phosphatase required for proper root hair development in Arabidopsis thaliana. Plant Cell 2008; 20:381-95; PMID:18281508; http://dx.doi.org/10.1105/tpc.107.054304
