Abstract
Many plant growth and developmental processes are modulated by the hormone auxin. Auxin-modulated proteolysis of Aux/IAAs, a family of transcriptional repressors, represents a major mode of auxin action. Auxin facilitates the interaction of Aux/IAAs with TIR1/AFB F-box proteins, promoting their ubiquitination by the SCFTIR1/AFB ubiquitin E3 ligase leading to subsequent degradation by the 26S proteasome. To identify new genes regulating Aux/IAA proteolysis in Arabidopsis thaliana, we took a genetic approach, identifying individuals with altered degradation of an IAA1-luciferase fusion protein (IAA1-LUC). A mutant with 2-fold slower IAA1-LUC degradation rate compared with wild-type was isolated. Positional cloning identified the mutant as an allele of TOPOISOMERASE6B, named top6b-7. TOP6B encodes a subunit of a plant and archea-specific enzyme regulating endoreduplication, DNA damage repair and transcription in plants. T-DNA insertion alleles (top6b-8 and top6b-9) were also analyzed. top6b-7 seedlings are less sensitive to exogenous auxin than wild-type siblings in primary root growth assays, and experiments with DR5:GUS. Additionally, top6b-7 seedlings have a 40% reduction in the amount of endogenous IAA. These data suggest that increased IAA1-LUC half-life in top6b-7 probably results from a combination of both lower endogenous IAA levels and reduced sensitivity to auxin.
Abbreviations
| LUC | = | luciferase |
| top6b | = | TOPOISOMERASE6B |
| 2 | = | 4-D, 2, 4-dichlorophenoxyacetic acid |
| IAA1-LUC | = | IAA1-luciferase fusion protein |
| IAA | = | indole-3-acetic acid |
| UPS | = | ubiquitin proteasome system |
Introduction
Auxins, a class of phytohormone typified by the major endogenous auxin indole-3-acetic acid (IAA), regulate many aspects of plant growth and development. Polar localization of auxin transporters in plasma membranes and localized auxin biosynthesis together establish auxin gradients within plant organs and tissues.Citation1 Such gradients have developmental consequences for patterning and phyllotaxy of new leaf primordia within the shoot apical meristem, lateral root initiation from the pericycle, and the overall establishment of plant organ polarity.Citation2 How such gradients are established and maintained over the developmental history of the plant and how cells sense and transduce differential auxin concentrations have been areas of intense study for several decades. By taking forward genetic, molecular genetic, and biochemical approaches many molecular components of auxin transport and signal transduction have been discovered.Citation3
Exogenous application of auxin rapidly modulates the transcription of a suite of genes, and this regulation is mediated through two families of transcription factors.Citation4 Auxin/IAA (Aux/IAA) proteins act as short-lived transcriptional regulators, of which there are 29 members in Arabidopsis thaliana.Citation5 These nuclear-localized proteins interact with and affect the activity of a family of transcription factors called Auxin Response Factors (ARFs), whose gene family size is 23 members in Arabidopsis.Citation6,7 The current model of auxin-mediated transcription proposes that when relieved from interaction with Aux/IAA proteins, ARFs modulate transcription of primary response genes that function in establishment and maintenance of patterning in plant organs.Citation5 A recent report describing the interaction of an Aux/IAA with a heat shock transcription factor in sunflower embryos suggests that Aux/IAA may regulate ARF independent processes, at least for one auxin-regulated response.Citation8
Our understanding of how auxin changes the activity of Aux/IAA proteins developed from the observation that auxin application stimulates rapid ubiquitin-mediated degradation of several endogenous Aux/IAA proteins or Aux/IAA fusion proteins in transgenic plants.Citation9-11 This increased Aux/IAA degradation rate is proposed to reduce the extent of Aux/IAA-ARF interactions, affecting transcription mediated by ARF proteins. However, not all Aux/IAA family members have equivalent degradation rates under high auxin conditions or show auxin-regulated proteolysis,Citation12 suggesting that auxin's effect on composition and concentration of Aux/IAA proteins is complex. Auxin modulates Aux/IAA ubiquitination by directly facilitating interaction of Aux/IAA proteins with the TIR1 family of F-box proteins and conversely, Aux/IAA proteins stabilize auxin's interaction with the F box protein.Citation13-15 F-box proteins are the substrate specificity subunits of SCF-type ubiquitin E3 ligases, a multiple subunit complex that interacts with the substrate and an E2 carrying activated ubiquitin to facilitate substrate ubiquitination. Thus, auxin-dependent enhancement of Aux/IAA interaction with its ubiquitin ligase results in Aux/IAA ubiquitination and subsequent degradation by the proteasome.Citation16 Recent work on the affinity of the TIR1/AFB proteins with multiple IAA proteins in vitro indicate that there are quantitative differences in their interactions, providing more evidence in support of a complex model for auxin transcriptional control.Citation17
In an effort to identify other trans-acting factors affecting Aux/IAA degradation, we treated a previously characterized transgenic Arabidopsis line expressing IAA1-LUCCitation11,12, Citation18-20 with a chemical mutagen (EMS) and screened for progeny with higher IAA1-LUC steady-state levels compared with the progenitor line.Citation21 We report here on one mutant recovered in this screen that has a 2-fold slower IAA1-LUC degradation rate. Surprisingly, this mutation mapped to the TOPOISOMERASE6B (TOP6B, At3g20780) locus.
TOP6B encodes the B subunit of the plant homolog of the archeabacterial DNA topoisomerase VI (Topo VI), a member of the B subfamily of type II topoisomerases. Type II topoisomerases catalyze ATP-dependent double-strand cleavage and decatenation, relieving tangling and supercoiling of DNA double strands that occur during replication, transcription, and recombination [for a review see refs.Citation22,23]. Archeabacterial Topo VI, and presumably others in the same subfamily, is an A2B2 heterotetramer.Citation24 The A subunits catalyze the double-stranded DNA cleavage reaction.Citation25,26 The B subunits contain a GHKL-type (gyrase, Hsp90, histidine kinase, MutL) ATPase domain,Citation24,27 a transducer domain, a helix–turn–helix domain, and a C-terminal domain.Citation26,28 The transducer domain links the B subunit to the A subunit and also contains a critical lysyl residue that contacts the γ-phosphate of ATP bound in the GHKL-domain.Citation26,28,29 Direct B-B subunit interaction occurs upon nucleotide binding and is thought to trap the DNA duplexes in the enzyme prior to strand cleavage and passage.
While type IIA topoisomerases can be found across all domains of life including some viruses, the topoisomerase IIB subfamily containing Topo VI homologs was initially thought to be archea-specific. The A subunit is present in eubacteria and metazoa as sporulation-specific protein 11 (SPO11); however, these organisms do not have any obvious B subunit homologs. Surprisingly, genes encoding both A and B subunits were identified in plants. Three and five genes encoding homologs of SPO11 were identified in Arabidopsis and rice, respectively, as well as a single copy of the B subunit, TOP6B, in these organisms.Citation30-32 AtTOP6B interacted with both AtSPO11–2 and AtSPO11–3 but not AtSPO11–1 in yeast-2-hybrid assay, suggesting that plants, unlike other eukaryotes, possess a topoisomerase VI composed of AtTOP6B and SPO11–2/SPO11–3.Citation32 Genetic evidence indicates that SPO11–1 and SPO11–2 function in meiotic recombination, while SPO11–3 appears to have a distinct function.Citation33 A number of mutants in these subunits were later identified by forward and reverse genetics giving some insight into the function of TOP6 in plants. Mutants in the A and B subunits were identified in screens searching for root hairless (RHL),Citation34-36 short dark-grown hypocotyls (HYP),Citation35,36 and brassinosteroid insensitive (BIN) mutants.Citation37 SPO11–3/BIN5/RHL2 and RHL3/BIN3/HYP6 encode the TOP6A and TOP6B subunits, respectively. Loss of function mutations in either subunit result in a severe pleiotropic phenotype: extreme dwarfism, lack of both root hairs and trichomes, small cells, photomorphogenesis in the dark, and brassinosteroid insensitivity. TOP6 mutants are defective in endoreduplication, as TOP6 appears to be required for production of a DNA content beyond 8C.Citation35,36,38
In addition to the core A2B2 subunits conserved in the TOP6 subclass of topoisomerases, AtTOP6 appears to include two additional plant-specific subunits required for enzyme activity. These subunits were identified from genetic screens as BIN4/MID1 and RHL1.Citation35, Citation39-41 The phenotypes of bin4/mid, rhl1, top6a/spo11–3 and top6b are indistinguishable from one another, suggesting that together they constitute a functional TOP6 in plants.
All together, the current data indicate that the phenotype of TOP6 loss of function plants is complex and results from a number of defects, including defects in endoreduplication, activation of a DNA damage response pathway, a reduction in heterochromatin, and the mis-expression of many genes. The characterization of bin4/mid revealed that TOP6B enzyme complex is not only required for endoreduplication, but also for heterochromatin formation during interphase and loss of TOP6 results in the activation of DNA damage response pathway.Citation40,42 A microarray analysis of both top6b/bin3 and top6a/bin5 mutants indicated mis-regulation of a large number of genes, including those involved in brassinosteroid and auxin signaling pathways.Citation37 Here, we report the isolation and characterization of a new TOP6B allele based on slowed IAA1-LUC degradation, and demonstrate that loss of TOP6 activity results in an auxin deficiency that affects responses to exogenous auxin.
Results
A genetic screen for mutants in Arabidopsis thaliana with altered IAA1-LUC degradation uncovers a new TOPOISOMERASE6B allele
To uncover novel regulators of Aux/IAA proteolysis, we developed a genetic screen to identify Arabidopsis thaliana mutants with altered Aux/IAA degradation by measuring the abundance and degradation rate of an IAA1-LUC fusion protein expressed from a transgene in individuals from a mutagenized population.Citation21 Our hypothesis was that seedlings with higher steady-state LUC activity than the progenitor line could have increased LUC activity from slower proteolysis of the fusion protein. Degradation of specific endogenous Aux/IAA proteins is difficult to measure due to their low abundance and presence of many related family members. IAA-LUC fusion proteins allow sensitive and quantitative determination of proteolytic rates. We previously showed that multiple Aux/IAA-LUC fusion proteins display basal degradation rates and auxin-modulated and proteasome inhibitor-sensitive degradation rates in vivo identical to tested endogenous Aux/IAA proteins.Citation11,12,18,19 Using this screen, we reported on the isolation of a novel viable allele of CULLIN1, a subunit of the E3 ligase responsible for Aux/IAA ubiquitination, demonstrating that the screen can successfully identify genes regulating Aux/IAA degradation.Citation21
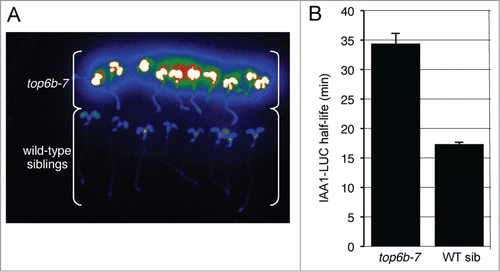
Additional screening identified a distinct M2 seedling with a higher IAA1-LUC steady-state level than other seedlings on the plate (, arrow). F1 backcross to the progenitor line gave normal LUC activity, indicating the mutation was recessive. F2 seedlings from the first backcross (BC1) segregated 3:1 for the mutant morphological phenotype, and all mutant seedlings had higher IAA1-LUC steady-state levels compared with wild type siblings indicating that the high LUC activity and the phenotype (see below) co-segregated (). To confirm that this mutant was a bona fide degradation mutant, we measured the IAA1-LUC degradation rate in multiple individuals from a BC2F2 population.Citation21 The half-life of IAA1-LUC in mutant seedlings averaged 34.5 min, statistically slower than the half-life (17.4 min) determined for IAA1-LUC in wild-type siblings (), confirming this mutant has impaired IAA1-LUC degradation.
Figure 1. top6b-7 has higher levels of IAA1-LUC and a slower IAA1-LUC degradation rate. (A) Confirmation of higher IAA1-LUC steady-state levels and co-segregation with the mutant phenotype. An F2 population of seeds from a backcross to the progenitor transgenic IAA1-LUC line was re-screened for the accumulation IAA1-LUC. Seedlings with the top6b phenotype (see ) were separated from wild type seedlings and imaged after pre-incubation with the substrate luciferin. (B) IAA1-LUC degradation rates in top6b-7 and wild type sibling seedlings. Using a single-seedling degradation assayCitation21 IAA1-LUC half-lives (t1/2) were determined in 7 d-old individual mutant and wild-type siblings in an BC2F2 segregating population. In top6b-7 seedlings IAA1-LUC t1/2 = 34.5 min, n = 59. In wild-type siblings IAA1-LUC t1/2 = 17.4 min, n = 180. Bars are standard error. Half-lives are statistically different by a Student's t test (P = 1.16*10−14, α = 0.05).
To positionally clone the mutant gene, we made a mapping population by out-crossing an M3 plant to Landsberg erecta (Ler). Mutant F2 plants from this cross were used as the mapping population. Additionally, we tested this population for LUC activity, and LUC activity segregated independently from the mutant phenotype indicating that the IAA1-LUC transgene was not linked to the mutation causing the mutant phenotype. Additionally, the 2-fold increase in half-life of IAA1-LUC only occurred in seedlings with a mutant phenotype, indicating that the longer half-life is not due to a cis mutation in the IAA1-LUC transgene. Bulked segregant analysisCitation43,44 linked the mutation to markers on the long arm of chromosome III (Fig. S2). Using a small mapping population (, Experiment 1), we were able to map the mutation between SNPs PERL0464591 and PERL0476408, and using a larger mapping population, we mapped the mutation between SNPs PERL0472202 and PERL0473223 (). This final mapping interval spanned ∼138 kb, and contained 35 open reading frames (www.arabidopsis.org).
Figure 2. Mapping of top6b-7 and identification of the mutation. (A) Positional cloning of top6b-7. For experiment 1, a small mapping population was used to roughly map, and a larger mapping population was used for experiment 2 fine mapping. Markers for mapping are shown with red tick marks. The mutation mapped between PERL0472202 and PERL0473223 SNPS. (B) Identification of mutation in top6b-7. Genomic DNA for At3g20780 was sequenced from 340 bp 5′ of START codon to 270 bp 3′ of the STOP codon. Sequence is shown for exon 1, intron 1, and exon 2 with intron sequence denoted by lowercase type. The G to A mutation in top6b-7 is bold underlined type. (C) Amino acid substitution in top6b-7. The first 100 amino acids of the TOP6B primary sequence are shown. The E36K substitution in top6b-7 is bold underlined type. (D) Multiple sequence alignment of TOP6B N-terminus. TOP6B sequences were retrieved from NCBI, by protein BLAST search using the Arabidopsis protein. Sequences were aligned using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/), and the sequences near the N-terminus are shown. An arrow indicates the glutamate residue changed to a lysine residue in top6b-7. Symbols for organisms are: Pp, Physcomitrella patens; Sm, Selaginella moellendorffii; Vv, Vitis vinifera; Rc, Ricinus communis; At, Arabidopsis thaliana; Sb, Sorghum bicolor; Os, Oryza sativa; Ps, Picea sitchensis; Cr, Chlamydomonas reinhardtii; Ta, Thermosphaera aggregans and; Si, Sulfolobus islandicus. Ta and Si are included to show divergence of plant TOP6B proteins from those of Archaea. (E) Gene structure of TOP6B and allele locations used in this study. The point mutation in top6b-7 is denoted by a small, black triangle. T-DNA insertions are indicated with large, black triangles with allele designated above them. Exons, UTRs, introns, and deletions are indicated by black boxes, white boxes, lines, and a small triangle, respectively. Scale bars represent 100 bp. Graphics were generated with Exon-Intron Graphic Maker (http://wormweb.org/exonintron).
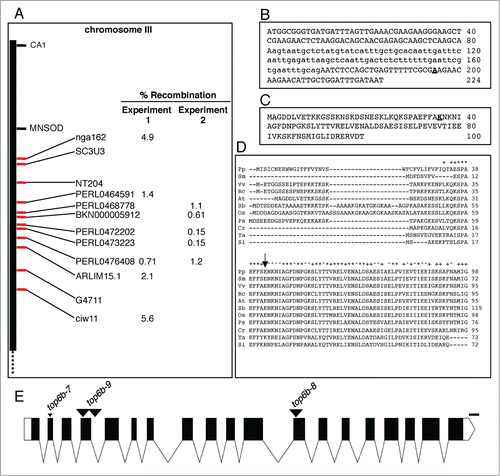
Next, we selected candidate genes in this region to sequence based on gene ontology annotations associated with the ubiquitin-proteasome system and also based on genes with similar mutant phenotypes. Mutations in TOPOISOMERASE6B (TOP6B, At3g20780.1) had been previously reported by several groups to result in extreme dwarfism,Citation34-38 similar to the phenotype of the mutant we recovered. We sequenced TOP6B in our mutant and found a G to A substitution in exon 2 () predicted to result in an E36K substitution (). This residue is conserved among the plants orthologs examined () and lies in a highly conserved region adjacent to the conserved “Motif B I” of the GHKL-type ATP binding domain.Citation45 Curiously, it is not conserved in the archeal proteins ().
Mutations in TOP6B Cause Severe Growth Defects
Mutations in TOP6B had been identified in previous genetic screens and in studies to characterize TOP6B in plants. These characterized alleles and corresponding references are summarized in . In order to assign an allele designation for the mutation identified in this study, we assigned the other mutants a top6b allele number based on the order of publication (). We designated our allele top6b-7, and obtained two additional previously uncharacterized T-DNA alleles, which we named top6b-8 and top6b-9 (, ). To ensure that the T-DNA insertions in top6b-8 and top6b-9 were correctly annotated, we determined the T-DNA insertion site in these alleles by sequencing the T-DNA specific genotyping PCR fragment with the primer specific to the T-DNA left border. A T-DNA PCR product for top6b-8 could only be obtained with the reverse gene-specific primer suggesting only one insertion at this site, and sequencing this PCR product placed the insertion in exon 12 ( and Fig. S2). For top6b-9, a PCR product could be obtained using either the forward or the reverse gene-specific primer with the T-DNA primer, suggesting the presence of two tandem inverted T-DNA insertions. Sequencing these products revealed the junction of one of the left-borders in exon 4 and the other in intron 4 ( and Fig. S2) with 139 bases of TOP6B sequence separating the left-border junctions. The presence of the sequence between the insertions, however, is uncertain as the right-border junctions were not sequenced.
Table 1. Alleles of TOP6B
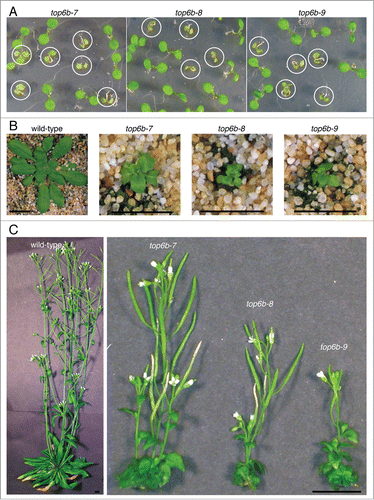
The phenotypes of top6b-7, -8, and -9 plants are all very similar. Plants homozygous for these three alleles are phenotypically indistinguishable from one another when germinated on agar plates. top6b seedlings are small compared with their wild-type siblings and have pale, epinastic, unexpanded cotyledons (). By 5 weeks, the growth defects in top6b plants are even more striking. The plants are extremely dwarfed with rosettes measuring less than one centimeter in diameter (). All three alleles have bolted and flowered by 10 weeks of growth (). The inflorescences of top6b plants are very short compared with those of wild-type plants, but flowers are fertile and produce siliques with a few seeds.
Figure 3. Morphological phenotype of top6b mutants. (A) top6b seedling phenotype. A population of seeds segregating for each allele was plated, cold-treated for 24 h at 4°C, and grown under continuous light for 10 d. Plants with the homozygous phenotype are circled. Seedlings not circled are wild-type segregants. (B) top6b mutant phenotype after 5 wk growth. Seeds segregating for each allele were sown directly on soil, cold-treated for 48 h at 4 °C, then grown 7 d at 16°C under constant. Seedling were selected by phenotype, and transplanted to individual pots for continued growth at 16°C. Scale bars are 1 cm. (C) top6b phenotype after 10 wk of growth. Plants in (B) were grown for an additional 5 wk as described above. Scale bars are 1 cm.
Top6b-7 is Allelic to T-DNA Insertion Alleles of TOP6B
To confirm that the mutation in top6b-7 was responsible for the observed phenotypes, we crossed top6b-7 to the T-DNA alleles for an allelism test. All three alleles segregate 3:1 for a wild-type to mutant phenotypic ratio (wt:m) when a heterozygous parent is selfed, indicating that the three alleles are recessive (). We crossed TOP6B/top6b-7 IAA1-LUC hmz plants in both directions to TOP6B/top6b-8 and TOP6B/top6b-9 plants and analyzed segregation of mutant phenotypes in the F1 progeny from these crosses. If the mutations are allelic, then the F1 progeny from the crosses should segregate 3wt:1 min. If not allelic, then all the F1 progeny should have a wild-type phenotype. The F1 progeny from all crosses tested segregated 3:1, indicated by a Chi-square test (), demonstrating that the mutation in top6b-7 is responsible for the observed morphological phenotype. To confirm that the F1 mutant individuals were heteroallelic and did not result from a self, several were genotyped for top6b-7 and the respective T-DNA allele and all tested were heteroallelic (data not shown).
Table 2. Allelism tests of top6b-7 morphological phenotype
To verify that the IAA1-LUC degradation defect is dependent on loss of TOP6B, the IAA1-LUC degradation rate was determined in F1 heteroallelic seedlings. If the degradation defect resulted from a mutation in a gene closely linked to TOP6B in top6b-7 and not from top6b-7 itself, then given the recessive nature of the decreased LUC degradation rate, none of the resulting heteroalleleic F1 plants from the allelism test crosses would have the longer half-life characteristic of top6b-7. The IAA1-LUC half-lives in mutant F1 individuals from all crosses were very similar to that calculated for top6b-7 mutants (), indicating that a defect in TOP6B is responsible for the longer IAA1-LUC half-life. Longer half-lives were only observed in seedlings with mutant phenotypes. F1 wild-type seedlings had a shorter half-life similar to that previously calculated for IAA1-LUC in wild-type Col and similar to that calculated in the F2 wild-type siblings from the self of TOP6B/top6b-7 BC2F1 (). All together, these data indicated that the mutation in top6b-7 is responsible for both the growth defects and for slower IAA1-LUC degradation observed in this line.
Table 3. Allelism test for IAA1-LUC degradation defects
Top6b-7 has Altered Auxin Physiology
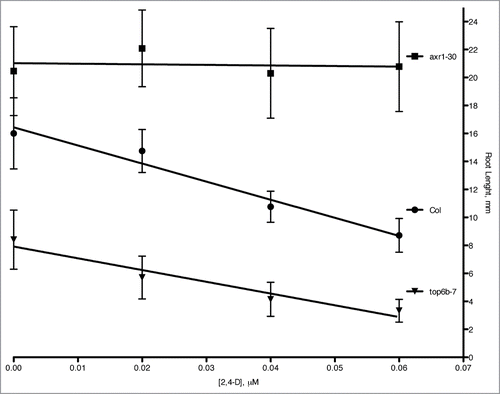
Because Aux/IAA degradation rates are affected by auxin levels and auxin responsiveness, we undertook experiments to examine if top6b-7 mutants had altered auxin physiology/responses. A classic and well-documented response to exogenous auxin is inhibition of primary root growth in seedlings. We tested the dose-responsiveness of top6b-7 primary roots to growth inhibition by application of the synthetic auxin, 2,4-dichlorophenoxyacetic acid (2,4-D) (). The auxin response of roots from a known auxin-resistant mutant, axr1–30 Citation21,46 grown at the same time was included. Plants were plated and grown continuously on the indicated concentration of 2,4-D, and primary root lengths were measured after 7 d of growth. As expected, axr1–30 roots are resistant to root growth inhibition at these concentrations of 2,4-D. Wild-type Col-0 roots also respond as predicted, with primary roots growing less on increasing concentrations of 2,4-D. The Col-0 line is statistically different from axr1–30 by a factorial ANOVA (p-value <0.001), and by comparing slopes axr1–30 is 97% less responsive to auxin than Col-0. The auxin response for top6b-7 is statistically different for Col-0 by a factorial ANOVA (p-value <0.001). By comparing the slope of the dose response curve in this analysis, top6b-7 is 35% less responsive to 2,4-D than Col-0, indicating that response to exogenous auxin is affected in top6b-7 seedlings.
Figure 4. Root growth inhibition in top6b-7 mutants with auxin treatment. Seeds were plated and grown on solid GM agar plates supplemented with the indicated concentration of 2,4-D for 7 d at 20°C with continuous lighting. The results are the combined data from three independent experiments. Values are mean ± 1 SD 59 ≤ n ≤ 68. The equations for linear regression are: y = −4.11(x) + 21.02, R2 = 0.0172; y = −129.32(x) + 16.44, R2 = 0.964; y = −83.98(x) + 7.91, R2 = 0.940 for axr1–30, Col, and top6b-7, respectively.
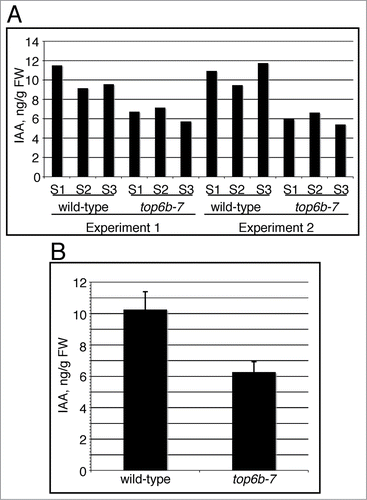
To determine whether auxin levels were affected in top6b-7, which could affect auxin response, we used LC-MS/MS to measure IAA levels from a segregating population of BC2 F2 seedlings. top6b-7 and wild-type seedlings were pooled into three samples each for two independent growth experiments, totaling six samples for each genotype. Wild-type seedlings have more IAA than their top6b-7 siblings in every sample measured (). Combining the six replicates, IAA levels averaged 10.26 and 6.26 ng/g FW in wild-type and top6b-7 seedlings, respectively (), and these measurements are statistically different by a Student's t test (p-value = 5.8·10−5, α = 0.05). IAA levels are therefore reduced approximately 40% in top6–7 seedlings.
Figure 5. IAA levels in top6b-7 mutants. Seeds segregating for top6b-7, were plated on GM, cold treated for 48 h at 4°C, and grown under constant light (50 μmol·m−1·s−2) at 20°C. Mutants and wild-type siblings were collected and sent for IAA measurement using LC-MS/MS. (A) IAA measurements are represented from 2 independent growth experiments. For each growth experiment, 3 different biological samples (S1-S3) for each phenotypic class were used for measurements. (B) The mean for the 6 replicates shown in (A) for each phenotypic class is shown, and are 10.39 and 6.26 ng IAA/g FW for top6b-7 and wild-type respectively. Bars are 1 sd. Averages are statistically different by a Student's t-Test (P = 5.84031*10−5, α = 0.05).
To further investigate endogenous auxin levels and auxin responsiveness in top6b-7, we crossed TOP6B/top6b-7 to a DR5:GUS reporter line (a minimal 35S promoter with synthetic auxin response elements driving expression of β-glucuronidase)Citation47 which has been shown to be an excellent sensor of endogenous auxin production in cells and tissues and also is very responsive to the application of exogenous auxin. We examined GUS staining patterns in a population of DR5:GUS seedlings segregating for top6b-7 with and without treatment with exogenous auxin (). Overall, the GUS staining pattern in top6b-7 mock-treated is similar to that of its wild-type siblings, with high GUS activity concentrated in the root apical meristem (white arrowheads), sites of lateral root emergence, and at the cotyledon apex. GUS activity, however, is reduced in top6b-7 shoot apical meristem (black arrowheads), in newly emerging leaves, and in the veins and peripheral cells of cotyledons (insets) (, row 1).
Figure 6. DR5:GUS expression and responsiveness to 2,4-D in top6b-7. Seeds from a TOP6B/top6b-7 DR5:GUS plants were grown for 10 d on solid GM plates at 22°C, under constant light. Seedlings were then transferred to liquid GM and treated with either solvent or 50 μM 2,4-D for 24 h at 22°C, under constant light. Seedlings were stained for GUS activity for 24 h and 1 h for mock and 2,4-D samples respectively. Two representative seedlings are shown for each sample. Black arrowheads denote shoot apices, and white arrowheads denote the root apical meristems. Insets are enlargements of cotyledons to show GUS staining pattern.
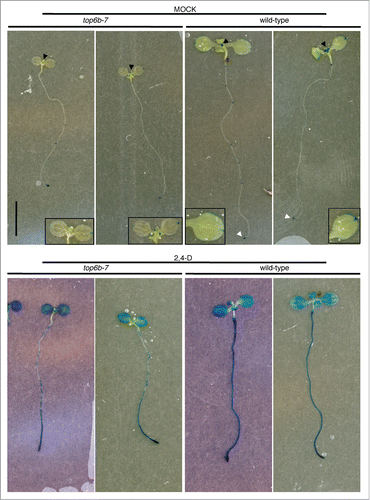
To examine DR5:GUS responses to exogenous auxin, we treated seedlings with 2,4-D and stained for GUS activity (, row 2). top6b-7 roots and true leaves are less sensitive to exogenous auxin in inducing GUS activity. All cells of wild-type roots have a continuous staining pattern and are darkly stained; however, under the same staining conditions, the roots of top6b-7 consist of patches of cells that do not stain or stain much less. Additionally, the true leaves of top6b-7 appear less sensitive to exogenous auxin than those of wild-type, with less intense staining. These results support our previous observations and are consistent with lowered endogenous auxin levels affecting responsiveness to exogenous auxin.
Discussion
Proteolysis by the ubiquitin proteasome system (UPS) is a major mode of regulation for plant growth, development, reproduction and responses to the environment. Genome sequencing and annotation efforts have revealed that components of the UPS are greatly expanded in plants compared with other eukaryotes, and in the model plant Arabidopsis thaliana an estimated 6% of genes are predicted to encode UPS components [for a review see ref. Citation48]. Well-characterized substrates of the UPS in plants are the Aux/IAA proteins, whose degradation is facilitated by, and required for, response to the phytohormone auxin. The current model is that Aux/IAAs are ubiquitinated by the E3 ubiquitin ligase SCFTIR1/AFB, and this modification targets them for degradation by the proteasome. Interaction with SCFTIR1/AFB requires the binding of auxin in the presence of both TIR1/AFB and the Aux/IAA to facilitate/stabilize the interaction. Mutations in a specific region of the Aux/IAAs, termed the degron, specifically core residues (GWPPV/L/I) of Aux/IAAs, slows proteolysis likely by disrupting their interactions with auxin and TIR1/AFB.Citation9,49,50 The core degron, however, does not confer the same degradation rate on all Aux/IAA family members,Citation12 leading to the hypothesis that other unknown factors might be modulating their degradation. Toward the goal of understanding the requirements for Aux/IAA degradation in Arabidopsis, we undertook a forward genetic screen to find mutants with slower degradation of an IAA1-LUC fusion protein.Citation21 Here we describe one mutant, top6b-7, isolated from this screen and present evidence that links topoisomerase VI to IAA levels and response.
Previous characterization of topoisomerase VI mutants in Arabidopsis revealed that plant growth requires the activity of this topoisomerase. One of the prominent molecular defects in these mutants is a block of endoreduplication. Endoreduplication, an alternative cell cycle, results in increased ploidy (called endoploidy) by replication of nuclear DNA without subsequent nuclear and cell divisions.Citation39,51,52 Plant cells undergo extensive endoreduplication as they differentiate and increase in size, and the extent of endoreduplication depends on the species and cell type. In Arabidopsis, cells range from 2C to 32C.Citation53 From analysis of Arabidopsis top6 mutants, this topoisomerase is thought to be specific to endoreduplication and the resolution of topological problems. The loss of TOP6 activity could make it physically impossible to further replicate DNA past 8C, with tangling becoming too extensive for replication to proceed, triggering the DNA damage response pathway that has been reported to be activated in top6 plants.Citation40,42
The molecular mechanisms of how increased ploidy contributes to cellular growth/expansion remain to be resolved, but it is generally believed that an increase in ploidy allows for greater metabolic activity of larger cells. This idea is supported by evidence that plant biotrophs induce endoreduplication at sites of nutrient exchange to boost the metabolic output of infected cells [for a review see ref. Citation54]. Many plant hormones including ethylene, gibberellic acid, auxins, brassinosteroids, and cytokinins regulate growth and promote cellular expansion, providing an explanation why mutants in components of the TOP6 complex were isolated in a screen for brassinosteroid insensitive mutants, and shown to be slightly resistant to exogenous brassinosteroid in hypocotyl elongation assays.Citation37,42 Microarray analysis of bin3/top6b and bin5/top6a also revealed these mutants are defective in brassinosteroid-induced gene expression and that many components of both auxin and brassinosteroid signaling pathways are under-expressed.Citation37
Brassinosteroid and auxin signaling pathways are interdependent to promote cell elongation, and both pathways must be functional in order for either hormone to induce growth.Citation55 Specifically, ARF2 is required for brassinosteroid responses.Citation56 Brassinosteroid application has been reported to stimulate expression of DR5:GUS without altering endogenous IAA levelsCitation57 indicating the gene targets of the two hormones’ signaling pathways have significant overlap, which confounds interpretation of auxin responsiveness and DR5:GUS expression in top6b-7. DR5:GUS expression in the roots of top6b-7 appeared to be less responsive to exogenous 2,4-D with patches of cells having very light staining to no staining. This result might be explained by mis-regulation of auxin transporters in top6b-7 roots, as brassinosteroid appears to influence expression of the PIN-FORMED (PIN) auxin transporters.Citation58 However, slower IAA1-LUC degradation in the mutant reveals an additional auxin-specific defect, because only auxin stimulates Aux/IAA degradation.Citation12,55
Examination of endogenous IAA levels in top6b-7 revealed a nearly 40% reduction compared with wild-type siblings, indicating that slower IAA1-LUC degradation could in large part result from lower auxin levels. Why IAA levels in top6b-7 are reduced remains an open question, but microarray analysis of bin3/top6b and bin5/top6a/spo1–3 indicated that YUCCA3 expression was ∼3–5-fold reduced in those lines,Citation37 suggesting an effect on auxin synthesis. IAA biosynthetic pathways are complex, not fully defined, and may be tryptophan (Trp)-dependent or –independent.Citation59 However, a major pathway for IAA synthesis is a two-step process from Trp to IAA via indole-3-pyruvate, with YUCCA family of proteins catalyzing the last stepCitation60 [for a review see refs.Citation59, Citation61, Citation62]. The YUCCA-dependent step is thought to be rate limiting for IAA synthesis since YUCCA overexpression lines have increased IAA and conjugated IAA, while overexpression of the penultimate enzyme has little effect on auxin levels.Citation60,63 However, the role of YUCCA3 specifically in regulating IAA levels is yet not known.
The phenotype of top6b is complex, and not simply a result of lower free IAA levels, as the growth defects cannot be rescued by application of auxin or brassinosteroid.Citation37 The increase in IAA1-LUC half-life in the mutant, most probably results from a combination of both a decreased endogenous IAA level and reduced sensitivity to auxin. Root growth inhibition assays with 2,4-D () demonstrate reduced sensitivity, which is also supported by the DR5:GUS staining in auxin treated top6b seedlings ( row 2). Basal DR5:GUS staining is also reduced in top6b seedlings, a result that could be explained by reduced sensitivity and/or reduced endogenous IAA levels. We, therefore, are left to conclude that a reduction in both sensitivity and IAA levels both contribute to the stabilization of IAA1-LUC in top6b-7. These mutants also have an activated DNA damage check-pointCitation42 and TOP6b also appears to function in an ROS response pathway binding to promoters of ROS-activated genes,Citation64 suggesting direct regulation of gene activity and processes that arrest growth are activated in top6 mutants.
In conclusion, the data presented here demonstrate for the first time the perturbation of auxin physiology in plants with disrupted topoisomerase VI activity as evidenced by 1) slower degradation of IAA1-LUC, 2) decreased levels of endogenous IAA, 3) reduced basal expression of DR5:GUS, and 4) reduced response to exogenous auxin in both root elongation assays and using the auxin responsive promoter DR5:GUS in top6b-7. The mechanism of IAA reduction remains an unanswered question, but it is tempting to speculate that reduced endoploidy of top6 cells decreases expression of IAA biosynthesis genes from either a reduction in copy number or by altering their transcriptional regulation. Alternatively, the activated DNA damage checkpoint in top6 plants could result in low IAA levels as a means to slow/arrest growth until the damage is repaired. The experiments described here provide evidence for a new pathway regulated by TOP6 activity.
Materials and Methods
Plant materials and growth conditions
All plants used in this study were Arabidopsis thaliana, ecotype Columbia-0 (Col) unless stated otherwise. Two T-DNA insertion lines for TOP6B, At3g20780, were obtained from the Arabidopsis Biological Resource Center (ABRC, http://abrc.osu.edu/) and named top6b-8 (SALK_024455c) and top6b-9 (SALK_10704)Citation65 [see ]. top6b-7 plants used for phenotypic analysis were all F2 plants from a second backcross to the progenitor line and all harbor the IAA1-LUC transgene (see genetic screen for details). All soil-grown plants were first cold-treated for 1–2 d, then grown under continuous white light at 16°C. Plants grown on solid agar growth media (GM)Citation21 were first surface sterilized as seeds in 30% bleach, 0.1% Trition-X 100 and cold-treated 1–2 d, then grown at 20°C under continuous white light (40–50 μmol·m−2·s−1) for the times indicated.
Genetic screen
The genetic screen for IAA1-LUC degradation mutants has been described previously.Citation21 In brief, a transgenic line expressing IAA1-LUC was treated with EMS, and M2 plants were screened for higher luciferase activity using a CCD camera (Princeton Instruments model NTE/CCd-TKD D12990). For this mutant, the higher luciferase activity was confirmed after the first backcross to the progenitor line (BC1 F2) using a different CCD camera (Andor Technology model DU 434-BV, Andor SOLIS software, South Windsor, CT). The BC1 F2 and BC2 F2 generations were used in the single seedling assay (described below). In these experiments, the degradation defect co-segregated with a phenotype, which was later used as a marker for following the mutation.
Positional cloning
A mapping population was generated by crossing an M3 plant to the Landsberg erecta (Ler) ecotype, and F1 plants from this cross were selfed to obtain F2 individuals. F2 individuals with the top6b parental phenotype were sacrificed for genomic DNA isolation. Bulk segregant analysisCitation44 on pooled DNA from approximately 50 mutant F2 individuals used a series of molecular markers that span the genome.Citation43 From this population, the mutation was linked to simple sequence length polymorphism (SSLP) marker nga162 (www.arabidopsis.org) on chromosome III. All individuals were then genotyped for additional markers that span chromosome III (experiment 1, ). Markers consisted of previously reported and annotated SSLP or cleaved amplified polymorphic sequence (CAPS) markers,Citation43,66 and CAPS markers derived from annotated single nucleotide polymorphisms (SNPs). SNPs that could be used as CAPS or dCAPS markers were determined using dCAPS finder 2.0 (http://helix.wustl.edu/dcaps/dcaps.html),Citation67 and new markers used in this study are listed in . For experiment 1, DNAs from a total of 71 mutant individuals were tested. Because of problematic genomic DNA preparations, not all markers worked for every individual. 71 individuals were genotyped for nga162, but only 37 individuals for marker ciw11. Individuals that were recombinant with nga162 and ciw11 markers were then genotyped for additional markers spanning the interval between them to narrow the mapping interval. For experiment 2, genomic DNA was prepared from 342 additional individuals. These individuals were genotyped in the interval between PERL0464591 and PERL0476408 with 329 individuals and 336 DNAs working for PERL0464591 and PERL0476408, respectively. Individuals recombinant at these markers, were then genotyped for additional markers within the interval. The mapping interval was narrowed to 138 kb between PERL0472202 and PERL0473223, and candidate genes in this interval were sequenced.
Genotyping top6b alleles
The mutation in top6b-7 creates an NruI restriction site that is missing in the wild-type sequence. We designed primers (5′TAACCCCAGCAACCAATCTC3′ and 5′GTTCCCGATC-GATAAGACCA3′) for PCR that amplify a region of 724 bp spanning the mutation. The PCR product was then digested with NruI and resolved by agarose gel electrophoresis. PCR and digestion of wild-type DNA results in one 724 bp band, top6–7 DNA results in 2 bands of 344 and 380 bp, and TOP6B/top6b-7 DNA results in three bands of 344, 380, and 724 bp. To genotype the T-DNA alleles top6b-8 and top6b-9, we used the gene-specific primers 5′GCCAAATGGCTGTCATTA-CTC3′ and 5′AAAGGATGACCTTCGAAGACC3′ for top6b-8 and 5′TTACCATCTTTGCACCCAGAC3′ and 5′TCGTGA-ATTTGCAGAATCTCC3′ for top6b-9. To genotype for the T-DNA insertion in these lines, we used a primer specific to the left border of the T-DNA, 5′TGGTTCACGTAGTGGGCCATC-G3′ with either gene-specific primer for each allele to test the direction of the insertion. Both gene-specific primers for top6b-9 when used with the T-DNA primer produced a DNA fragment, suggesting two tandem T-DNA insertions in this line. To verify the position of the T-DNA in these alleles, T-DNA-specific PCR products for each allele were sequenced using the T-DNA primer.
IAA1-LUC degradation experiments
BC2 F2 seeds segregating for top6b-7 were plated and grown in 96-well flat-bottom plates (Whatman, Clifton, NJ) with 100 μL solid GM for 7 d under constant light at 22°C. The experiments followed a protocol previously describedCitation21 using cycloheximide assays. Light emission from seedlings was measured every 15 min over a 60 min time course. To calculate IAA1-LUC half-lives in individual seedlings, the RLU (relative light units) at each time-point was normalized to the zero time-point measurement. To linearize the data, the ln(normalized RLU) was calculated. These data (y-axis) were then plotted against their respective time-points (x-axis), and the slope of this line determined for each seedling in the plate. The following formula was used to calculate the half-life: half-life (min) = (slope)/ln(0.5). Seedlings were scored as either mutant or wild-type based on the morphological phenotype, and long half-lives always occurred in seedlings that were visibly different from wild-type. The average half-life from multiple seedlings with wild-type or mutant phenotypes was then calculated for the final respective half-life calculation for each phenotype.
Allelism test
To determine if the mutation identified in top6b-7 was responsible for the observed morphological phenotype and IAA1-LUC degradation defect, we crossed top6b-7 to plants heterozygous for T-DNA alleles top6b-8 and top6b-9. F1 progeny from these crosses were plated on GM plates and grown 10 d under constant light at 20°C, then scored for segregation of the mutant phenotype. Some individuals with mutant phenotypes were sacrificed for genomic DNA extraction and genotyping for top6b using the strategy described above.
To determine if F1 mutant individuals that originated from the crosses also had slower IAA1-LUC half-lives, single-seedling degradation assays performed on those individuals. Seedlings were transferred from the GM plates into 96-well plates, treated with 100 μl of 1 mM D-luciferin in liquid GM solvent for 1 h in the dark. After the 1 h incubation, cycloheximide (2 mg/mL stock dissolved in water) was added to final concentration of 200 μg/ml. Light emission from seedlings was measured every 15 min over a 60 min time-course and half-lives calculated as described above.
Auxin response experiments
Seeds for axr1–30, Col, and TOP6B/top6b-7 were surface-sterilized, and grown on 2,4-dichlorophenoxyacetic acid (2,4-D, Sigma) plates at the indicated concentration. All media including the “0” 2,4-D contained the same percentage of solvent as a control. Seedlings were grown at 20°C under constant light for 7 d, then transferred to the surface of agar plates, imaged with flat-bed scanner, and roots measured with Image J v1.36b (http://rsbweb.nih.gov/ij/). Three independent experiments were performed using different batches of 2,4-D plates for each replicate.
IAA measurement
A population of seeds segregating for top6b-7 were plated on solid GM plates, and cold-stratified at 4 °C for 48 h, then grown for 10 d at 20°C under constant light (50 μmol·m−2·s−1). Seedlings from multiple plates were pooled together (50–100 mg) into three different samples for each phenotype, wild-type and top6b-7. Seedlings were grown for two independent experiments, with three samples for each phenotype per experiment. For both experiments, tissue was collected mid-morning to control for circadian regulation of IAA biosynthesis. Tissue was flash-frozen in N2 and sent for IAA quantification at the proteomics and mass spectrometry facility at the Donald Danforth Plant Science Center, St. Louis, MO USA (http://www.danforthcenter.org/science/core_facilities/proteomics_and_mass_spectrometry/). Samples were quantified by LC-MS/MS (20 min run) with d5-IAA as an internal standard.Citation68
DR5:GUS assays
To generate the plant lines used for these assays, pollen from a BC2 F2 top6b-7 homozygote was crossed to a previously published and well characterized DR5:GUS line.Citation47 F1 plants were genotyped for top6b-7 to ensure plants originated from the cross. The DR5:GUS T-DNA confers resistance to hygromycin, and F2 seedlings were selected for hygromycin resistance and transplanted to soil. F2 individuals were again genotyped for top6b-7, and F3 seedlings from top6b-7 F2 heterozygotes were tested for segregation of hygromycin resistance to select for lines homozygous for the DR5:GUS transgene. The IAA1-LUC T-DNA also carries a 35S:GUS expression transgene that confers strong constitutive GUS expression; therefore, we also examined the LUC activity in F3 seedlings segregating for top6b-7 and homozygous for the DR5:GUS T-DNA (by hygromycin resistance) to ensure that the IAA1-LUC transgene was absent in those lines. F3 lines segregating for top6b-7 and homozygous for DR5:GUS in which the IAA1-LUC transgene had segregated away were used for analysis.
For these assays, seedlings were grown on GM plates for 10 d at 22°C under continuous light. Seedlings were treated with 2,4-D as indicated prior to GUS staining. For GUS staining, tissue was washed twice (20 min) in GUS assay buffer (25 mM Na2HPO4/NaH2PO4 pH 7.0, 1.25 mM K3Fe(CN)6, 1.25 mM K4Fe(CN)6, 0.25% Triton-X 100, 0.25 mM EDTA, 20% methanol). After the second wash, the buffer was replaced with 2.4 mM X-Gluc (5-bromo-4-chloro-3-indoxyl-β-D-glucuronide cyclohexylammonium salt, stock dissolved in N,N-dimethylformamide) in GUS assay buffer and incubated at 37°C for the indicated times. After the staining reaction was complete, the seedlings were transferred to 100% ethanol to clear chlorophyll from the tissue.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors gratefully acknowledge support from the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, US. Department of Energy (contracts DE-FG02–03ER15416 and DE-FG02–09ER16077) to JC. In addition, the initial development of the genetic screen was supported in part by a grant from the National Science Foundation (IBN 0212659) to JC. Additional support was from the Paul K. and Ruth R. Stumpf Endowed Professorship in Plant Biochemistry to J. Callis. We wish to acknowledge the UC-Davis Controlled Environment Facility (CEF) for assistance in the propagation of transgenic plants. We would like to thank Joey Lasiter and Gurjeet Bath for help with positional cloning and genotyping, Tania Gonzalez for assistance with auxin dose-response experiments, and Marissa Simon for advice on GUS experiments. We thank former and current members of the Callis laboratory and Dr. Anne Britt (UC-Davis) for helpful discussions.
References
- Boutté Y, Ikeda Y, Grebe M. Mechanisms of auxin-dependent cell and tissue polarity. Curr Opin Plant Biol 2007; 10:616-23; PMID:17720615; http://dx.doi.org/10.1016/j.pbi.2007.07.008
- Vieten A, Sauer M, Brewer PB, Friml J. Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci 2007; 12:160-8; PMID:17369077; http://dx.doi.org/10.1016/j.tplants.2007.03.006
- Sauer M, Robert S, Kleine-Vehn J. Auxin: simply complicated. J Exp Bot 2013; 64:2565-77; PMID:23669571; http://dx.doi.org/10.1093/jxb/ert139
- Pierre-Jerome E, Moss BL, Nemhauser JL. Tuning the auxin transcriptional response. J Exp Bot 2013; 64:2557-63; PMID:23630231; http://dx.doi.org/10.1093/jxb/ert100
- Chapman EJ, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet 2009; 43:265-85; PMID:19686081; http://dx.doi.org/10.1146/annurev-genet-102108-134148
- Guilfoyle TJ, Hagen G. Auxin response factors. Plant Growth Regul 2001; 20:281-91.
- Remington DL, Vision TJ, Guilfoyle TJ, Reed JW. Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol 2004; 135:1738-52; PMID:15247399; http://dx.doi.org/10.1104/pp.104.039669
- Carranco R, Espinosa JM, Prieto-Dapena P, Almoguera C, Jordano J. Repression by an auxin/indole acetic acid protein connects auxin signaling with heat shock factor-mediated seed longevity. Proc Natl Acad Sci U S A 2010; 107:21908-13; PMID:21115822; http://dx.doi.org/10.1073/pnas.1014856107
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 2001; 414:271-6; PMID:11713520; http://dx.doi.org/10.1038/35104500
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 2001; 13:2809-22; PMID:11752389; http://dx.doi.org/10.1105/tpc.13.12.2809
- Zenser N, Ellsmore A, Leasure C, Callis J. Auxin modulates the degradation rate of Aux/IAA proteins. Proc Natl Acad Sci U S A 2001; 98:11795-800; PMID:11573012; http://dx.doi.org/10.1073/pnas.211312798
- Dreher KA, Brown J, Saw RE, Callis J. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 2006; 18:699-714; PMID:16489122; http://dx.doi.org/10.1105/tpc.105.039172
- Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature 2005; 435:441-5; PMID:15917797; http://dx.doi.org/10.1038/nature03543
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 2005; 9:109-19; PMID:15992545; http://dx.doi.org/10.1016/j.devcel.2005.05.014
- Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 2005; 435:446-51; PMID:15917798; http://dx.doi.org/10.1038/nature03542
- Maraschin Fdos S, Memelink J, Offringa R. Auxin-induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J 2009; 59:100-9; PMID:19309453; http://dx.doi.org/10.1111/j.1365-313X.2009.03854.x
- Calderón Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, et al. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 2012; 8:477-85; PMID:22466420; http://dx.doi.org/10.1038/nchembio.926
- Ramos JA, Zenser N, Leyser O, Callis J. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 2001; 13:2349-60; PMID:11595806; http://dx.doi.org/10.1105/tpc.13.10.2349
- Worley CK, Zenser N, Ramos J, Rouse D, Leyser O, Theologis A, Callis J. Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J 2000; 21:553-62; PMID:10758506; http://dx.doi.org/10.1046/j.1365-313x.2000.00703.x
- Zenser N, Dreher KA, Edwards SR, Callis J. Acceleration of Aux/IAA proteolysis is specific for auxin and independent of AXR1. Plant J 2003; 35:285-94; PMID:12887580; http://dx.doi.org/10.1046/j.1365-313X.2003.01801.x
- Gilkerson J, Hu J, Brown J, Jones A, Sun TP, Callis J. Isolation and characterization of cul1-7, a recessive allele of CULLIN1 that disrupts SCF function at the C terminus of CUL1 in Arabidopsis thaliana. Genetics 2009; 181:945-63; PMID:19114460; http://dx.doi.org/10.1534/genetics.108.097675
- Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem 2001; 70:369-413; PMID:11395412; http://dx.doi.org/10.1146/annurev.biochem.70.1.369
- Singh B, Sopory S, Reddy M. Plant DNA topoisomerases: structure, function, and cellular roles in plant development. Crit Rev Plant Sci 2004; 23:251-69; http://dx.doi.org/10.1080/07352680490452816
- Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 1997; 386:414-7; PMID:9121560; http://dx.doi.org/10.1038/386414a0
- Buhler C, Lebbink JH, Bocs C, Ladenstein R, Forterre P. DNA topoisomerase VI generates ATP-dependent double-strand breaks with two-nucleotide overhangs. J Biol Chem 2001; 276:37215-22; PMID:11485995; http://dx.doi.org/10.1074/jbc.M101823200
- Corbett KD, Benedetti P, Berger JM. Holoenzyme assembly and ATP-mediated conformational dynamics of topoisomerase VI. Nat Struct Mol Biol 2007; 14:611-9; PMID:17603498
- Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci 2000; 25:24-8; PMID:10637609; http://dx.doi.org/10.1016/S0968-0004(99)01503-0
- Corbett KD, Berger JM. Structure of the topoisomerase VI-B subunit: implications for type II topoisomerase mechanism and evolution. EMBO J 2003; 22:151-63; PMID:12505993; http://dx.doi.org/10.1093/emboj/cdg008
- Corbett KD, Berger JM. Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI. Structure 2005; 13:873-82; PMID:15939019; http://dx.doi.org/10.1016/j.str.2005.03.013
- Hartung F, Puchta H. Molecular characterisation of two paralogous SPO11 homologues in Arabidopsis thaliana. Nucleic Acids Res 2000; 28:1548-54; PMID:10710421; http://dx.doi.org/10.1093/nar/28.7.1548
- An XJ, Deng ZY, Wang T. OsSpo11-4, a rice homologue of the archaeal TopVIA protein, mediates double-strand DNA cleavage and interacts with OsTopVIB. PLoS One 2011; 6:e20327; PMID:21637817; http://dx.doi.org/10.1371/journal.pone.0020327
- Hartung F, Puchta H. Molecular characterization of homologues of both subunits A (SPO11) and B of the archaebacterial topoisomerase 6 in plants. Gene 2001; 271:81-6; PMID:11410368; http://dx.doi.org/10.1016/S0378-1119(01)00496-6
- Stacey NJ, Kuromori T, Azumi Y, Roberts G, Breuer C, Wada T, Maxwell A, Roberts K, Sugimoto-Shirasu K. Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J 2006; 48:206-16; PMID:17018031; http://dx.doi.org/10.1111/j.1365-313X.2006.02867.x
- Schneider K, Wells B, Dolan L, Roberts K. Structural and genetic analysis of epidermal cell differentiation in Arabidopsis primary roots. Development 1997; 124:1789-98; PMID:9165126
- Sugimoto-Shirasu K, Roberts GR, Stacey NJ, McCann MC, Maxwell A, Roberts K. RHL1 is an essential component of the plant DNA topoisomerase VI complex and is required for ploidy-dependent cell growth. Proc Natl Acad Sci U S A 2005; 102:18736-41; PMID:16339310; http://dx.doi.org/10.1073/pnas.0505883102
- Sugimoto-Shirasu K, Stacey NJ, Corsar J, Roberts K, McCann MC. DNA topoisomerase VI is essential for endoreduplication in Arabidopsis. Curr Biol 2002; 12:1782-6; PMID:12401175; http://dx.doi.org/10.1016/S0960-9822(02)01198-3
- Yin Y, Cheong H, Friedrichsen D, Zhao Y, Hu J, Mora-Garcia S, Chory J. A crucial role for the putative Arabidopsis topoisomerase VI in plant growth and development. Proc Natl Acad Sci U S A 2002; 99:10191-6; PMID:12119417; http://dx.doi.org/10.1073/pnas.152337599
- Hartung F, Angelis KJ, Meister A, Schubert I, Melzer M, Puchta H. An archaebacterial topoisomerase homolog not present in other eukaryotes is indispensable for cell proliferation of plants. Curr Biol 2002; 12:1787-91; PMID:12401176; http://dx.doi.org/10.1016/S0960-9822(02)01218-6
- Breuer C, Ishida T, Sugimoto K. Developmental control of endocycles and cell growth in plants. Curr Opin Plant Biol 2010; 13:654-60; PMID:21094078; http://dx.doi.org/10.1016/j.pbi.2010.10.006
- Kirik V, Schrader A, Uhrig JF, Hulskamp M. MIDGET unravels functions of the Arabidopsis topoisomerase VI complex in DNA endoreduplication, chromatin condensation, and transcriptional silencing. Plant Cell 2007; 19:3100-10; PMID:17951446; http://dx.doi.org/10.1105/tpc.107.054361
- Schneider K, Mathur J, Boudonck K, Wells B, Dolan L, Roberts K. The ROOT HAIRLESS 1 gene encodes a nuclear protein required for root hair initiation in Arabidopsis. Genes Dev 1998; 12:2013-21; PMID:9649505; http://dx.doi.org/10.1101/gad.12.13.2013
- Breuer C, Stacey NJ, West CE, Zhao Y, Chory J, Tsukaya H, Azumi Y, Maxwell A, Roberts K, Sugimoto-Shirasu K. BIN4, a novel component of the plant DNA topoisomerase VI complex, is required for endoreduplication in Arabidopsis. Plant Cell 2007; 19:3655-68; PMID:18055605; http://dx.doi.org/10.1105/tpc.107.054833
- Lukowitz W, Gillmor CS, Scheible WR. Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol 2000; 123:795-805; PMID:10889228; http://dx.doi.org/10.1104/pp.123.3.795
- Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci U S A 1991; 88:9828-32; PMID:1682921; http://dx.doi.org/10.1073/pnas.88.21.9828
- Malik SB, Ramesh MA, Hulstrand AM, Logsdon JM Jr. Protist homologs of the meiotic Spo11 gene and topoisomerase VI reveal an evolutionary history of gene duplication and lineage-specific loss. Mol Biol Evol 2007; 24:2827-41; PMID:17921483; http://dx.doi.org/10.1093/molbev/msm217
- Hotton SK, Eigenheer RA, Castro MF, Bostick M, Callis J. AXR1-ECR1 and AXL1-ECR1 heterodimeric RUB-activating enzymes diverge in function in Arabidopsis thaliana. Plant Mol Biol 2011; 75:515-26; PMID:21311953; http://dx.doi.org/10.1007/s11103-011-9750-8
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 1997; 9:1963-71; PMID:9401121; http://dx.doi.org/10.1105/tpc.9.11.1963
- Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 2009; 10:385-97; PMID:19424292; http://dx.doi.org/10.1038/nrm2688
- Tian Q, Nagpal P, Reed JW. Regulation of Arabidopsis SHY2/IAA3 protein turnover. Plant J 2003; 36:643-51; PMID:14617065; http://dx.doi.org/10.1046/j.1365-313X.2003.01909.x
- Yang X, Lee S, So JH, Dharmasiri S, Dharmasiri N, Ge L, Jensen C, Hangarter R, Hobbie L, Estelle M. The IAA1 protein is encoded by AXR5 and is a substrate of SCF(TIR1). Plant J 2004; 40:772-82; PMID:15546359; http://dx.doi.org/10.1111/j.1365-313X.2004.02254.x
- Sugimoto-Shirasu K, Roberts K. “Big it up”: endoreduplication and cell-size control in plants. Curr Opin Plant Biol 2003; 6:544-53; PMID:14611952; http://dx.doi.org/10.1016/j.pbi.2003.09.009
- Mizukami Y. A matter of size: developmental control of organ size in plants. Curr Opin Plant Biol 2001; 4:533-9; PMID:11641070; http://dx.doi.org/10.1016/S1369-5266(00)00212-0
- Galbraith DW, Harkins KR, Knapp S. Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol 1991; 96:985-9; PMID:16668285; http://dx.doi.org/10.1104/pp.96.3.985
- Wildermuth MC. Modulation of host nuclear ploidy: a common plant biotroph mechanism. Curr Opin Plant Biol 2010; 13:449-58; PMID:20542725; http://dx.doi.org/10.1016/j.pbi.2010.05.005
- Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2004; 2:E258; PMID:15328536; http://dx.doi.org/10.1371/journal.pbio.0020258
- Vert G, Walcher CL, Chory J, Nemhauser JL. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci U S A 2008; 105:9829-34; PMID:18599455; http://dx.doi.org/10.1073/pnas.0803996105
- Nakamura A, Higuchi K, Goda H, Fujiwara MT, Sawa S, Koshiba T, Shimada Y, Yoshida S. Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol 2003; 133:1843-53; PMID:14605219; http://dx.doi.org/10.1104/pp.103.030031
- Li L, Xu J, Xu Z-H, Xue H-W. Brassinosteroids stimulate plant tropisms through modulation of polar auxin transport in Brassica and Arabidopsis. Plant Cell 2005; 17:2738-53; PMID:16141452; http://dx.doi.org/10.1105/tpc.105.034397
- Korasick DA, Enders TA, Strader LC. Auxin biosynthesis and storage forms. J Exp Bot 2013; 64:2541-55; PMID:23580748; http://dx.doi.org/10.1093/jxb/ert080
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci U S A 2011; 108:18512-7; PMID:22025724; http://dx.doi.org/10.1073/pnas.1108434108
- Mano Y, Nemoto K. The pathway of auxin biosynthesis in plants. J Exp Bot 2012; 63:2853-72; PMID:22447967; http://dx.doi.org/10.1093/jxb/ers091
- Zhao Y. Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol Plant 2012; 5:334-8; PMID:22155950; http://dx.doi.org/10.1093/mp/ssr104
- Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T. Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 2007; 143:1362-71; PMID:17220367; http://dx.doi.org/10.1104/pp.106.091561
- Simková K, Moreau F, Pawlak P, Vriet C, Baruah A, Alexandre C, Hennig L, Apel K, Laloi C. Integration of stress-related and reactive oxygen species-mediated signals by Topoisomerase VI in Arabidopsis thaliana. Proc Natl Acad Sci U S A 2012; 109:16360-5; PMID:22988090; http://dx.doi.org/10.1073/pnas.1202041109
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003; 301:653-7; PMID:12893945; http://dx.doi.org/10.1126/science.1086391
- Kwon M, Lee K, Choe S. Novel Simple Sequence Length Polymorphic (SSLP) Markers for Postional Cloning in Arabidopsis thaliana. Korean J of Genetics 2005; 27:1-8.
- Neff MM, Turk E, Kalishman M. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet 2002; 18:613-5; PMID:12446140; http://dx.doi.org/10.1016/S0168-9525(02)02820-2
- Pan X, Welti R, Wang X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat Protoc 2010; 5:986-92; PMID:20448544; http://dx.doi.org/10.1038/nprot.2010.37
