Abstract
Leaf morphogenesis requires the establishment of adaxial-abaxial polarity in emerging leaf primordia, and a number of genes participating in this process have been identified in recent years. We previously reported that the 26S proteasome is important in specifying the leaf adaxial fate. More recently, two papers from separate researches showed that several genes encoding ribosomal large subunit proteins also play an important role in leaf adaxial-abaxial patterning. Here we show that plants with a single mutation in the genes encoding either 26S proteasome subunits or ribosomal proteins shared similar abnormalities in some leaves, with an outgrowth formed on the distal part of the leaf abaxial side. Plants harboring these 26S proteasome or ribosome mutations in combination with an additional mutation asymmetric leaves1 or 2 (as1 or as2) demonstrated severely defective leaves, and the phenotypes of these double mutants were very similar. Because activities of the 26S proteasome and ribosome both affect the level of functional proteins, the recent findings suggest that a previously unrecognized regulation, the protein level regulation, is critical in normal leaf patterning. A regulatory model for the 26S proteasome and ribosome actions in leaf patterning is discussed.
Addendum to: Yao Y, Ling Q, Wang H, Huang H. Ribosomal proteins promote leaf adaxial identity. Development 2008; 135:1325-34.
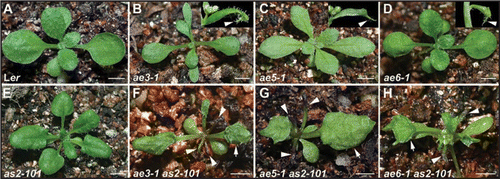
Recently, several Arabidopsis genes encoding the 26S proteasome subunit and ribosomal proteins have been identified that play important roles in specifying leaf adaxial identity.Citation1–Citation4 The 26S proteasome and ribosome are large protein or protein/rRNA complex, and mutations in different protein genes of each complex could result in plants with leaf adaxial-abaxial defects, whereas the phenotypes are relatively weak. To explore whether or how these two complexes may cooperate in controlling leaf patterning, we characterized genetically three genes among others of the complexes: AE3, AE5 and AE6, which encode a 26S proteasome subunit RPN8A, and ribosomal proteins RPL28A and RPL5A, respectively. Compared with wild-type Ler plants (), ae3-1 (), ae5-1 () and ae6-1 () did not exhibited strong leaf polarity defects,Citation1–Citation3 but some leaves from each of these three mutants produced an outgrowth on the distal part of the adaxial leaf side (, insets). In contrast to their single mutants and as2-101 (), double mutants containing as2–101 or as1–101 and ae3-1, ae5-1 or ae6-1 resulted in plants with severe but similar leaf phenotypes (, for the as1 combinations, data not shown). Briefly, most leaves were radially symmetric (, arrowheads) and the remaining expanded lotus-like leaves had very rough adaxial surfaces (, arrows).
It was previously proposed that several proteins from the 26S proteasome or ribosome complex demonstrated specific functions distinct from those of their complexes in protein degradation or translation.Citation5–Citation8 However, mutations in different 26S proteasome or ribosomal protein genes examined all resulted in a similar leaf defect, albeit varying in severity, and double mutants with as1 or as2 all produced strong and very similar leaf phenotypes.Citation1,Citation2,Citation4 Based on these observations, it seems unlikely that the regulation of leaf patterning depends on functions of a particular protein of the complexes, but instead, the conserved functions of protein degradation or translation of the two complexes may be involved. How these two complexes function to determine leaf polarity is not yet clear. One possibility is that these two systems are required for an accurate balance in levels between the adaxial- and abaxial-promoting factors during leaf polarity formation. These factors include transcriptional factors and proteins required for small RNA biogenesis and action (reviewed in refs. Citation9 and Citation10), as two microRNAs, miR165 and miR166, and one trans-acting siRNA, tasiR-ARF, are important in leaf patterning.Citation11–Citation14 It is known that some of the regulatory factors for leaf patterning act antagonistically, and exhibit complementary expression domains in multiple tissues. For example, adaxial-promoting genes REV/PHB/PHV antagonize abaxial-promoting ones KAN1/KAN2/KAN3Citation15,Citation16 and tasiR-ARF which specifies the adaxial leaf fate antagonizes abaxial-promoters miR165/miR166.Citation17 On the other hand, the 26S proteasome and ribosome complexes are known to act selectively to process their targets,Citation18,Citation19 and certain leaf patterning factors are likely to be the targets of these complexes. Therefore, a failure in degrading an abaxial-promoting factor (a loss of function in the 26S proteasome) and incapability in synthesizing its corresponding antagonistic adaxial-promoting factor (a loss of function in ribosome) can result in the same consequence for leaf patterning. This model can explain why ae3, ae5 and ae6 single mutants share some similar abnormalities in the leaf and the severe leaf phenotypes of ae3 as2, ae5 as2 and ae6 as2 double mutants are very similar with each other.
Figures and Tables
Figure 1 Mutant phenotypes suggest that the protein-level regulation is critical for normal leaf patterning. (A–E) Phenotypes of wild-type and single mutants. (A) wild-type Ler, (B) ae3-1, (C) ae5-1, (D) ae6-1 and (E) ae2–101. Insets in (B–D) show cauline leaves with an ectopic outgrowth (arrowheads) on their distal part of the abaxial side. (F–H) Double mutant phenotypes of ae3-1 as2–101 (F), ae5-1 as2–101 (G) and ae6-1 as2–101 (H). Arrowheads and arrows in (F–H) show the radially symmetric and lotus-like leaves with rough adaxial leaf surfaces, respectively. Bars = 5 mm in (A–H).
Acknowledgements
This research was supported by grants from the Chinese National Scientific Foundation (30630041 and 30721061), and Chinese Academy of Sciences (KSCX2-YW-N-016) to H.H.
Addendum to:
References
- Yao Y, Ling Q, Wang H, Huang H. Ribosomal proteins promote leaf adaxial identity. Development 2008; 135:1325 - 1334
- Pinon V, Etchells JP, Rossignol P, Collier SA, Arroyo JM, Martienssen RA, Byrne ME. Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development 2008; 135:1315 - 1324
- Huang W, Huang H. A novel function of the 26S proteasome in repressing class-1 KNOX genes during leaf development. Plant Signaling and Behavior 2007; 2(1):25 - 27
- Huang W, Pi L, Liang W, Xu B, Wang H, Cai R, Huang H. The proteolytic function of the Arabidopsis 26S proteasome is required for specifying leaf adaxial identity. Plant Cell 2006; 18:2479 - 2492
- Cho YH, Yoo SD, Sheen J. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 2006; 127:579 - 589
- Nishimura T, Wada T, Yamamoto KT, Okada K. The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell 2005; 17:2940 - 2953
- Smalle J, Kurepa J, Yang P, Babiychuk E, Kushnir S, Durski A, Vierstra RD. Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 2002; 14:17 - 32
- Weijers D, Franke-van Dijk M, Vencken RJ, Quint A, Hooykaas P, Offringa R. An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 2001; 128:4289 - 4299
- Xu L, Yang L, Huang H. Transcriptional, post-transcriptional and post-translational regulations of gene expression during leaf polarity formation. Cell Res 2007; 17:512 - 519
- Kidner CA, Timmermans MC. Mixing and matching pathways in leaf polarity. Curr Opin Plant Biol 2007; 10:13 - 20
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005; 121:207 - 221
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 2004; 18:2368 - 2379
- Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev 2003; 17:49 - 63
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell 2002; 110:513 - 520
- Hawker NP, Bowman JL. Roles for class III HD-Zip and KANADI genes in Arabidopsis root development. Plant Physiol 2004; 135:2261 - 2270
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 2003; 13:1768 - 1774
- Nogueira FT, Madi S, Chitwood DH, Juarez MT, Timmermans MC. Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev 2007; 21:750 - 755
- Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell 2007; 131:557 - 571
- Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 2004; 55:555 - 590