Abstract
A major challenge in biology is unravelling the mechanisms that regulate gene expression and an important task in this challenge is to identify binding sites for transcription factors in gene promoter regions. These binding sites are short DNA segments that are called motifs. Recent advances in genome sequence availability and in high-throughput gene expression analysis technologies have allowed for the development of computational methods for motif finding. The aim of our study was to identify NO-responsive elements in the promoter region of NO-regulated genes, based on microarray analyses. Using Genomatix Gene2Promotor and MatInspector, we identified eight families of TFBS occurred at least 15% more often in the promoter regions of the responsive genes in comparison to the promoter regions of 28447 Arabidopsis control genes. Most of these TFBSs, such as ocs element-like sequences and WRKY, were already reported to be involved in particular stresses response.
Addendum to: Palmieri MC, Sell S, Huang X, Scherf M, Werner T, Durner J, Lindermayr C. Nitric oxide-responsive genes and promoters in Arabidopsis thaliana: a bioinformatics approach. J Exp Bot 2008; 59:177-86.
Nitric oxide (NO) is an important signalling molecule in plants, which is implicated in many different physiological functions, such as disease resistance, stomata closure, seed germination, iron homeostasis, different development processes and response to abiotic stresses.Citation1–Citation4 NO mainly regulates these physiological processes by direct interaction with enzymes resulting in altered activities or by affecting gene transcription.
But how can NO regulate gene expression? The main idea in gene expression is that every gene contains the information to produce a protein. Gene expression begins with binding of protein factors, known as transcription factors, to enhancer and promoter sequences and transcription factors regulate the gene expression by activating or inhibiting the transcription machinery. It is assumed that co-expression of genes arises mainly from transcriptional co-regulation. As co-regulated genes are known to share some similarities in their regulatory mechanism, their promoter regions might contain some common motifs that are binding sites for transcription factors. A sensible approach to detect these regulatory elements is to search for overrepresented motifs in the promoter region of such a set of co-expressed genes. An overrepresented motif means a motif that occurs more often than one would expect by chance.
The aim of our study was to identify NO-responsive elements in the promoter region of NO-regulated genes. Therefore we did a whole genome transcript analyses from NO treated plants and NO-donor treated cell cultures to search for co-regulated genes, since similar expression profiles might be caused by the coordinated action of transcription factors. 28 genes were upregulated both in plant and cell culture experiments. Furthermore, 121 genes and 79 genes were induced exclusively in plant and cell culture experiments, respectively. Additionally, 26 genes were found to be downregulated. To screen these sets of co-regulated genes for common transcription factors binding site (TFBS) we used the Genomatix software.Citation5 First, the regulatory regions of the NO-responsive genes were extracted and defined as 500 base pairs upstream and 100 base pairs downstream of the transcription start. The isolated 601 bp promoter regions were then screened for common transcription binding sites with Genomatix MatInspector. We identified eight TFBS families, which occurred at least 15% more often in the promoter regions of the analyzed groups of genes in comparison to the promoter regions of the control genes. As control we used the occurrence of the transcription binding sites within a set of 28447 Arabidopsis genes in percentage (Genomatix database, http://www.genomatix.de).
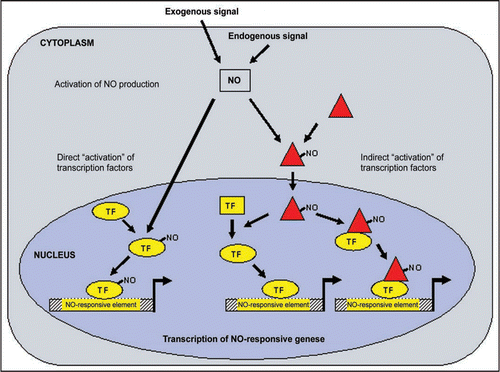
GBOX-, OCSE-and L1BX-elements are enriched in genes upregulated in both cell culture and plant experiments. Moreover, GBOX- and OCSE-elements, together with MYCL and OPAQ, are highly present in promoters of upregulated cell culture genes. In genes induced in plants only WRKY binding sites are enriched in their promoters. In the downregulated genes of cell cultures an increased occurrence of TBPF- and MIIG-elements was observed. We developed a general model illustrating the regulation of NO-responsive genes (). NO production is induced by exogenous or endogenous signals. The produced NO can react directly with a transcription factor resulting in its activation and binding to its specific NO-responsive element (see on the left side of the model).
In the animal system, Hausladen et al., (1996) showed how the transcription factor OxyR can process different redox-related signals into distinct transcriptional responses. By generating several stable, posttranslational modifications of the single regulatory thiol (SH), such as S-NO, S-OH and S-SG, they could shown that each modified forms were transcriptionally active but different in DNA binding affinity and promoter activities.Citation6
But a transcription factor can also be activated by another protein in a NO-dependent manner. After this protein is activated by nitric oxide, it can alter the conformation of a transcription factor or it can interact with a transcription factor to enable DNA-binding (see on the right side of the model). In both cases the transcription factor are activated indirectly by NO.
An example for indirect redox-dependent modification of transcription regulator is NPR1, a key regulator of systemic acquired resistance (SAR). During the SA-mediated activation of defence, there is a change in the cellular redox status that results in reduction of NPR1 to its active monomer form. In this form NPR1 is translocated into the nucleus where it interacts with the transcription factor TGA, the activator of SA-responsive elements in the promoter of PR genes.Citation7 Since thiol groups are involved in this regulatory process it is absolutely possible that NO can affect NPR1/TGA1 interaction and/or DNA-affinity of TGA1 by S-nitrosylation of regulatory thiol groups.
Taken together, such a bioinformatics approach is a useful tool to identified common transcription factor binding sites and promoter modules in co-regulated genes, which might be involved in establishing specific expression profiles. Since co-expression can be due to a variety of co-regulatory mechanisms, such analyses may be a fist step to provide the basis to understand regulatory networks involved in gene expression profiles. Next, the involvement of the identified motives in NO response should be experimentally validated.
Figures and Tables
Figure 1 Model illustrating the regulation of NO-responsive genes. NO production is induced by exogenous or endogenous signals. The produced NO can activate transcription factors directly or indirectly. In both cases, the transcription factors bind to its specific NO-responsive element, such as WRKY, GBOX, OCSE and/or OPAQ. TF: transcription factor.
Addendum to:
References
- Delledonne M, et al. Nitric oxide functions as a signal in plant disease resistance. Nature 1998; 394:585 - 588
- Durner J, Wendehenne D, Klessig DF. Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc Natl Acad Sci USA 1998; 95:10328 - 10333
- Garcia Mata C, Lamattina L. Nitric oxide and abscisic acid cross talk in guard cells. Plant Physiol 2002; 128:790 - 792
- Zhao L, et al. Nitric oxide functions as a signal in salt resistance in the calluses from two ecotypes of reed. Plant Physiol 2004; 134:849 - 857
- Quandt K, et al. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res 1995; 23:4878 - 4884
- Hausladen A, et al. Nitrosative stress: activaton of the transcription factor OxyR. Cell 1996; 86:719 - 729
- Pieterse CM, Van Loon LC. NPR1: the spider in the web of induced resistance signaling pathways. Curr Opin Plant Biol 2004; 7:456 - 464