Abstract
Jasmonates (JAs) control many aspects of plant defense and development, for instance by inhibiting growth and eliciting secondary metabolism. The mechanisms by which JAs regulate these processes are currently under intensive investigation. Examination of transcriptional changes upon methyl jasmonate (MeJA) perception in a fast-growing Arabidopsis thaliana cell suspension culture revealed a quick and direct dual effect of JAs on the plant\'s cellular processes. Simultaneously, JA-elicited Arabidopsis cells activated phenylpropanoid metabolism and repressed cell cycle progression. Early JA response genes were predominantly implicated in transcriptional regulation and JA biosynthesis. In two parallel screens, we identified both early responsive transcriptional activators (ORA47 and MYC2) and transcriptional repressors (STZ/ZAT10 and AZF2) that putatively regulate the expression of the JA biosynthesis gene LOX3. In this addendum, we provide additional data demonstrating that MYC2, STZ/ZAT10, and AZF2 might also control the expression of JAZ1/TIFY10a that encodes a key repressor in the JA signaling cascade.
Addendum to: Pauwels L, Morreel K, De Witte E, Lammertyn F, Van Montagu M, Boerjan W, Inzé D, Goossens A. Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc Natl Acad Sci USA 2008; 105:1380-5.
Jasmonates (JAs) are plant-specific signaling molecules that steer a diverse set of physiological and developmental processes and that are most nown for their ability to invoke defense responses, including the biosynthesis of secondary metabolites. This property has made JAs excellent tools to unravel biosynthetic pathways by combining JA elicitation with transcript and metabolite profiling.Citation1 In addition to activating defense pathways, JAs negatively regulate growthCitation2,Citation3 that can be correlated with the negative effect of JAs on cell cycle progression.Citation4 In our study,Citation5 we examined the effect of MeJA elicitation on the transcriptome of a cell suspension culture of Arabidopsis thaliana. MeJA induced the expression of genes involved in phenylpropanoid metabolism and associated pathways. Simultaneously, genes activated during the M-phase of the cell cycle were repressed. These results showed for the first time the transcriptional basis underlying the growth-to-defense transition activated by JAs.
The F-boxprotein CORONATINE INSENSITIVE 1 (COI1; At2g39940) plays a key role in JA signaling and loss of COI1 function leads to JA insensitivity.Citation2 Recently, the jasmonate ZIM-domain (JAZ) proteins, members of the tify protein family,Citation6 have been identified as the targets of COI1.Citation7,Citation8 In the presence of Ile-conjugated JA, JAZ proteins bind COI1, marking them for degradation by the 26S proteasome.Citation8 JAZ proteins function as repressors of JA signaling, probably by binding to the bHLH transcriptional activator MYC2 (At1g32640) and inhibiting its function.Citation7
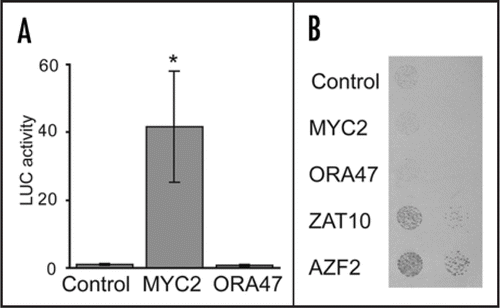
We reported that JAZ and MYC2 transcript levels rapidly increased in Arabidopsis cells after MeJA treatment.Citation5 In addition, early MeJA-induced genes encoded transcription factors from various protein families, as well as JA biosynthesis genes. To unravel the relation between these co-expressed genes, we set up two parallel screens. In one screen, transcription factors were transiently overexpressed in protoplastsCitation5 and the promoter activity of the JA biosynthesis gene LOX3 (At1g17420) was monitored. The results revealed that MYC2 could activate LOX3 expression, matching earlier observations that JA-induced LOX3 expression is reduced in the MYC2-defective mutant jin1.Citation9 In this addendum, we show that overexpressing MYC2 also activates the promoter of JAZ1/TIFY10a (At1g19180) (). This observation is in accordance with the induction of many JAZ genes in 35S:MYC2 plants and the ability of MYC2 to binding the G-box or its variant T/G-box in the promoter of JAZ3/TIFY6b (At3g17860), motifs also present in the JAZ1/TIFY10a promoter.Citation7
In addition to MYC2, we found that also the AP2/EREBP transcription factor ORA47 (At1g74930) functioned as a transcriptional activator and was capable of activating the promoter-driven expression of LOX3. ORA47 had been forwarded as a positive regulator of JA biosynthesis, whose action appeared to depend on COI1,Citation10 placing its function downstream of MYC2. Interestingly, contrary to that of MYC2, overexpression of ORA47 did not enhance the activation of the JAZ1/TIFY10a promoter, suggesting that ORA47 might regulate only a subset of the genes controlled by MYC2.
In the other screen, we assessed the binding potential of early induced transcription factors using a yeast one-hybrid approach.Citation5 In this manner, we identified two C2H2 zinc finger proteins, STZ/ZAT10 (At1g27730) and AZF2 (At3g19580) capable of binding the LOX3 promoter. STZ/ZAT10 and AZF2, as well as others of the 20-member C1-2i subfamily of C2H2 zinc finger proteins have been reported to be involved in plant stress responses.Citation11 Here, we show that both zinc finger proteins also bind the JAZ1/TIFY10a promoter in a similar yeast one-hybrid assay ().
Together our results illustrate that the complexity of the JA signaling cascade, in particular at the early responsive stage. Within this cascade, a central role is reserved for the JAZ/MYC2 interplay. Upon perception of the JA signal, JAZ repressors are removed and MYC2 released to activate downstream JA responses. In addition, ORA47 and possibly other, AP2/EREBP factors as activator(s), and C1-2i-type C2H2 zinc finger proteins as repressors, impinge on this central JAZ/MYC2 system in a coordinated fashion and guarantee fine-tuning or appropriate control of downstream gene expression. An intriguing subject for future studies will be to determine when and how all of these proteins connect and which additional signals or factors might affect this link. As for the MYC2 and JAZ proteins, the rapid induction of all of these genes might rather reflect an endpoint within the cascade rather than a beginning.
Figures and Tables
Figure 1 Identification of transcriptional regulators of JAZ1/TIFY10a expression. (A) Regulation of JAZ1/TIFY10a expression by transient expression of MYC2 and ORA47 open reading frames (ORF). Tobacco (Nicotiana tabacum) protoplasts were transfected with a PJAZ1:fLUC reporter construct, a P35S:ORF effector construct, and a P35S:rLUC normalization construct. The PJAZ1 construct covered a 1356-bp region preceding the start codon. Averaged (n ≥ 8) normalized fLUC activities are plotted relative to the P35S:GUS control. Error bars represent standard error. Asterisks indicate significant effects (t-test, p-value <0.05). (B) Yeast one-hybrid interaction with the JAZ1/TIFY10a promoter. Full-length ORFs fused to GAL4AD were expressed in a PJAZ1:HIS3 reporter strain. The empty Gateway destination vector was used as a negative control. Yeast was grown for 3 days on selective medium in presence of 3-AT. All methods are as described.Citation5
Addendum to:
References
- Goossens A, Häkkinen ST, Laakso I, Seppänen Laakso T, Biondi S, De Sutter V, Lammertyn F, Nuutila AM, Söderlund H, Zabeau M, Inzé D, Oksman Caldentey KM. A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc Natl Acad Sci USA 2003; 100:8595 - 8600
- Devoto A, Turner JG. Regulation of jasmonate-mediated plant responses in Arabidopsis. Ann Bot 2003; 92:329 - 337
- Yan Y, Stolz S, Chételat A, Reymond P, Pagni M, Dubugnon L, Farmer EE. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 2007; 19:2470 - 2483
- Swiatek A, Lenjou M, Van Bockstaele D, Inzé D, Van Onckelen H. Differential effect of jasmonic acid and abscisic acid on cell cycle progression in tobacco BY-2 cells. Plant Physiol 2002; 128:201 - 211
- Pauwels L, Morreel K, De Witte E, Lammertyn F, Van Montagu M, Boerjan W, Inzé D, Goossens A. Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc Natl Acad Sci USA 2008; 105:1380 - 1385
- Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G. The tify family previously known as ZIM. Trends Plant Sci 2007; 12:239 - 244
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García Casado G, López Vidriero I, Lozano FM, Ponce MR, Micol JL, Solano R. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007; 448:666 - 671
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 2007; 448:661 - 665
- Lorenzo O, Chico JM, Sánchez Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004; 16:1938 - 1950
- Pré MR. Functional analysis of jasmonate-responsive AP2/ERF-domain transcription factors in Arabidopsis thaliana 2006; The Netherlands University of Leiden Ph. D. Thesis
- Ciftci Yilmaz S, Mittler R. The zinc finger network of plants. Cell Mol Life Sci 2008; In Press http://dx.doi.org/10.1007/s00018-007-7473-4