Abstract
Seed germination initiates the postembryonic development of plants, which determines successful seedling establishment and plant propagation. It is therefore tightly regulated by diverse environmental conditions, including high salinity and drought, as well as by intrinsic developmental programs, among which gibberellic acid (GA) is best understood. Regulatory roles of GA in seed germination have been extensively studied. It is also known that high salinity inhibits germination by repressing genes encoding GA biosynthetic enzymes. However, it is still unclear how salt signals are coordinately incorporated into the GA signaling pathway at the molecular level. We recently demonstrated that a membrane-bound NAC transcription factor, NTL8, mediates salt signaling, primarily through a RGL2-independent GA pathway, in regulating seed germination. High salinity promotes NTL8 transcription and proteolytic activation of NTL8. Notably, the NTL8-mediated salt signaling is independent of abscisic acid (ABA). These observations indicate that membrane-mediated transcription control is an important component of salt signaling during seed germination.
Addendum to: Kim SG, Lee AK, Yoon HK, Park CM. A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J 2008; doi: 10.1111/j.1365-313X.2008.03493.x
Seed germination is an essential developmental event in plants because it determines the site of plant growth and thus successful plant propagation. Seeds constantly monitor environmental changes. Light, moisture and temperature are three main factors that regulate seed germination.Citation1,Citation2 The external signals are incorporated into internal developmental programs, among which GA and ABA have been most extensively studied. ABA and GA confer antagonistic effects on seed germination through complicated signaling crosstalks. The former delays seed germination, while the latter promotes seed germination.Citation3–Citation5 Acordingly, favorable environmental conditions activate genes encoding ABA deactivating enzymes and those encoding GA biosynthetic enzymes.Citation6
Even under favorable conditions, seed germination can be delayed by other factors, such as high salinity. It has been shown that high salinity delays seed germination by reducing GA biosynthesis.Citation7 ABA is a major plant growth hormone mediating plant responses to high salinity, suggesting that the salt effects on seed germination would depends on this growth hormone. We observed that the GA synthetic enzyme gene GA3ox1 is significantly repressed by high salinity. In particular, the salt repression of GA3ox1 is independent of ABA. The GA3ox1 transcription is efficiently repressed by high salinity even in the ABA-deficient aba3-1 mutant to a level comparable to that observed in control plants treated with high salt. These observations support that an ABA-independent salt signaling pathway is also involved in the GA regulation of seed germination.
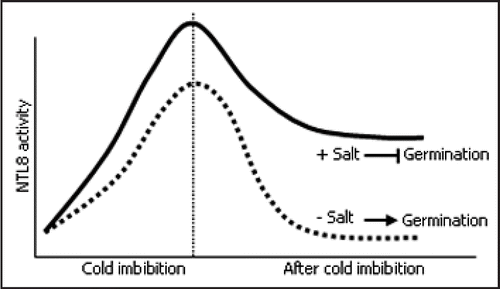
NTL8 is a membrane-bound NAC transcription factor in Arabidopsis.Citation8 It is unique among the NTL members in that the NTL8 transcription is induced by high salinity specifically in the cold-imbibed seed. While germination of control seeds is greatly reduced in the presence of high salt, that of the ntl8-1 mutant seeds is resistant to high salinity. In contrast, seeds of the transgenic plants overexpressing an active NTL8 form (NTL8ΔC) germinated very poorly under high salt conditions, indicating that NTL8 plays a role in the salt regulation of seed germination. However, the NTL8 transcription is unaffected by ABA, and germination of the ntl8-1 knockout mutant seeds is reduced by ABA to a degree similar to that of control seeds under the same conditions. It is therefore evident that NTL8-mediated salt signaling is independent of ABA.
It is notable that NTL8-mediated salt regulation of seed germination is intimately related to GA signaling. RGL2 is one of the DELLA proteins that negatively regulate GA signals.Citation9 When GA biosynthesis is induced, the RGL2 protein is degraded via the ubiquitin-dependent degradation pathway, causing promotion of seed germination.Citation10 The Arabidopsis GA biosynthetic mutant, ga1–3, does not germinate in the absence of exogenously applied GA, indicating that GA is essential for seed germination. In contrast, the ga1–3 rgl2 double mutant seeds germinate nearly 100% even in the absence of exogenous GA application, indicating that RGL2 is a negative regulator of GA function in seed germination.Citation9 However, recent studies strongly support that there would be additional negative regulators other than RGL2 in GA signaling.Citation11,Citation12
NTL8 is dramatically induced in the imbibed seed, and its induction is further elevated by cold treatment. However, the NTL8 induction decreases to a basal level within 24 hours after cold imbibition. This pattern is very similar to that of RGL2. Germination of the ntl8-1 mutant seeds is nearly 100% in the presence of 100 µM paclobutrazol (PAC), an inhibitor of GA synthesis. The NTL8 protein is released from the membranes through proteolytic cleavage. We found that this activation process is triggered in the PAC-treated plants. Together, these observations demonstrate that NTL8 is a negative regulator of GA signaling, particularly under high salt conditions.
While it is widely documented that environmental conditions greatly affect seed germination, only a few genes have been reported to mediate the environmental signals.Citation7,Citation13,Citation14 We found that NTL8 is one of such signaling mediators that are functioning in monitoring environmental fluctuations. Furthermore, NTL8 is essentially independent of RGL2, although some degree of feedback control was observed between them. It is envisioned that NTL8 may be a molecular link that integrates external signals into the intinsic developmental pathways during seed germination.
GA biosynthesis increases during cold imbibition.Citation15 However, seed germination is delayed at cold temperatures. In addition, germination is delayed or inhibited when seeds are completely waterlogged, suggesting that signaling network governing seed germination is more complicated than previously thought. Both the NTL8 and RGL2 genes are rapidly induced when seeds are cold-imbibed. It is assumed that the high levels of their transcripts would be maintained if the conditions are unfavorable, while they would quickly decrease under favorable conditions. It will be interesting to examine how environmental factors regulate their transcription and how the perceived signals are transmitted to the unidentified downstream genes. Extensive analysis of genome-wide expression profiling using the published microarray data would be helpful in identifying the signaling mediators.Citation17
A related question is how salt stress regulates GA biosynthesis. The previous and our data revealed that it suppresses the transcription of GA3ox1 encoding a GA biosynthetic enzyme, supporting that high salinity affects GA biosynthesis. The ABA-mediated salt stress reduces seed germination by imposing both osmotic stress and ion homeostasis. However, we found that the NTL8-mediatd salt signaling alters ion homeostasis but does not detectably impose osmotic stress. This observation is in contrast to the previous finding that high salinity imposes osmotic stress in inhibiting seed germination.Citation18,Citation19 One possible scenario is that the NTL8-mediated pathway is a specific salt signaling pathway that exerts its role only under high salinity, while the ABA-mediated salt signaling pathway plays a somewhat general role in seed germination under a variety of environmental stress conditions.
GA is the primary growth hormone that promotes seed germination, independently or cooperatively with other growth hormones, such as ABA and ethylene, through diverse signaling crosstalks. Notably, the NTL8-mediated salt signals, which have a negative regulatory effect on seed germination, are also incorporated into the germination-promoting GA signaling pathway. This finding provides a clue as to how the NTL8 pathway contributes to seed germination. It would not be a major signaling pathway that regulates seed germination. Instead, this signaling pathway would be required particularly under unfavorable conditions. In the presence of high salt, the germinating seeds are unable to complete their life cycle. We therefore conclude that the NTL8-mediated salt signaling pathway is required to inhibit seed germination under high salt conditions. NTL8 would be a molecular linkage that modulates seed germination by incorporating salt stress signals into the GA signaling cascades, providing an adaptive strategy to ensure that seeds are allowed to germinate only when the surrounding conditions are favorable ().
Figures and Tables
Figure 1 NTL8 regulation of GA-mediated salt signaling in seed germination. High salinity reduces GA biosynthesis. GA represses both NTL8 transcription and NTL8 processing. However, it is likely that the NTL8-mediated signaling is not required under favorable growth conditions. It inhibits seed germination only when the surrounding conditions are unfavorable for plant propagation. NTL8 may function as a molecular link that incorporates environmental signals into the GA-mediated signaling pathway in seed germination.
Addendum to:
References
- Finch Savage WE, Cadman CS, Toorop PE, Lynn JR, Hilhorst HW. Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J 2007; 51:60 - 78
- Penfield S, Josse EM, Kannangara R, Gilday AD, Halliday KJ, Graham IA. Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr Biol 2005; 15:1998 - 2006
- Koornneef M, Bentsink L, Hilhorst H. Seed dormancy and germination. Curr Opin Plant Biol 2002; 5:33 - 36
- Gubler F, Millar AA, Jacobsen JV. Dormancy release, ABA and pre-harvest sprouting. Curr Opin Plant Biol 2005; 8:183 - 187
- Peng J, Harberd NP. The role of GA-mediated signalling in the control of seed germination. Curr Opin Plant Biol 2002; 5:376 - 381
- Seo M, Hanada A, Kuwahara A, Endo A, Okamoto M, Yamauchi Y, North H, Marion Poll A, Sun TP, Koshiba T, Kamiya Y, Yamaguchi S, Nambara E. Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J 2006; 48:354 - 366
- Lopez Molina L, Mongrand S, Chua NH. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 2001; 98:4782 - 4787
- Kim SY, Kim SG, Kim YS, Seo PJ, Bae M, Yoon HK, Park CM. Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Res 2007; 35:203 - 213
- Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is upregulated following imbibition. Genes Dev 2002; 16:646 - 658
- Itoh H, Matsuoka M, Steber CM. A role for the ubiquitin-26S-proteasome pathway in gibberellin signaling. Trends Plant Sci 2003; 8:492 - 497
- Bassel GW, Zielinska E, Mullen RT, Bewley JD. Downregulation of DELLA genes is not essential for germination of tomato, soybean and Arabidopsis seeds. Plant Physiol 2004; 136:2782 - 2789
- Ariizumi T, Steber CM. Seed germination of GA-insensitive sleepy1 mutants does not require RGL2 protein disappearance in Arabidopsis. Plant Cell 2007; 19:791 - 804
- Oh E, Kim J, Park E, Kim JI, Kang C, Choi G. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 2004; 16:3045 - 3058
- Lee S, Lee EJ, Yang EJ, Lee JE, Park AR, Song WH, Park OK. Proteomic identification of annexins, calcium-dependent membrane binding proteins that mediate osmotic stress and abscisic acid signal transduction in Arabidopsis. Plant Cell 2004; 16:1378 - 1391
- Yamauchi Y, Ogawa M, Kuwahara A, Hanada A, Kamiya Y, Yamaguchi S. Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 2004; 16:67 - 78
- Dat JF, Capelli N, Folzer H, Bourgeade P, Badot PM. Sensing and signalling during plant flooding. Plant Physiol Biochem 2004; 42:273 - 282
- Becerra C, Puigdomenech P, Vicient CM. Computational and experimental analysis identifies Arabidopsis genes specifically expressed during early seed development. BMC Genomics 2006; 7:38
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, van der Straeten D, Peng J, Harberd NP. Integration of plant responses to environmentally activated phytohormonal signals. Science 2006; 311:91 - 94
- Khan MA, Gul B, Weber DJ. Action of plant growth regulators and salinity on seed germination of Ceratoides lanata. Can J Bot 2004; 82:37 - 42