Abstract
Flowering is a developmental process, which is influenced by chemical and environmental stimuli. Recently, our research established that the Arabidopsis SUMO E3 ligase, AtSIZ1, is a negative regulator of transition to flowering through mechanisms that reduce salicylic acid (SA) accumulation and involve SUMO modification of FLOWERING LOCUS D (FLD). FLD is an autonomous pathway determinant that represses the expression of FLOWERING LOCUS C (FLC), a floral repressor. This addendum postulates mechanisms by which SIZ1-mediated SUMO conjugation regulates SA accumulation and FLD activity.
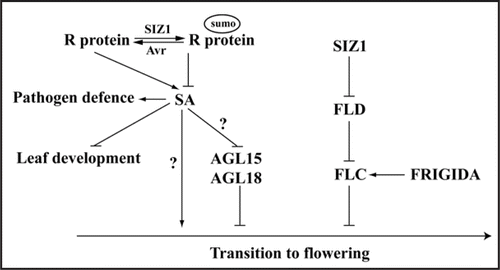
SUMO conjugation and deconjugation are post-translational processes implicated in plant defense against pathogens, abscisic acid (ABA) and phosphate (Pi) starvation signaling, development, and drought and temperature stress tolerance, albeit only a few of the modified proteins have been identified.Citation1–Citation8 The Arabidopsis AtSIZ1 locus encodes a SUMO E3 ligase that regulates floral transition and leaf development.Citation8,Citation9 siz1 plants accumulate substantial levels of SA, which is the primary cause for dwarfism and early short-day flowering exhibited by these plants.Citation1,Citation9 How SA promotes transition to flowering is not yet known but apparently, it is through a mechanism that is independent of the known floral signaling pathways.Citation9,Citation10 Exogenous SA reduces expression of AGAMOUS-like 15 (AGL15), a floral repressor that functions redundantly with AGL18.Citation11,Citation12 A possible mechanism by which SA promotes transition to flowering may be by repressing expression of AGL15 and AGL18 ().
siz1 mutations also cause constitutive induction of pathogenesis-related protein genes leading to enhanced resistance against biotrophic pathogens.Citation1 Several bacterial type III effector proteins, such as YopJ, XopD and AvrXv4, have SUMO isopeptidase activity.Citation13–Citation15 PopP2, a member of YopJ/AvrRxv bacterial type III effector protein family, physically interacts with the TIR-NBS-LRR type R protein RRS1, and possibly stabilizes the RRS1 protein.Citation16 Phytopathogen effector and plant R protein interactions lead to increased SA biosynthesis and accumulation, which in turn activates expression of pathogenesis-related proteins that facilitate plant defense.Citation17 SIZ1 may participate in SUMO conjugation of plant R proteins to regulate Avr and R protein interactions leading to SA accumulation, which, in turn, affects phenotypes such as diseases resistance, dwarfism and flowering time ().
Our recent work revealed also that AtSIZ1 facilitates FLC expression, negatively regulating flowering.Citation9 AtSIZ1 promotes FLC expression by repressing FLD activity.Citation9 Site-specific mutations that prevent SUMO1/2 conjugation to FLD result in enhanced activity of the protein to represses FLC expression, which is associated with reduced acetylation of histone 4 (H4) in FLC chromatin.Citation9 FLD, an Arabidopsis ortholog of Lysine-Specific Demethylase 1 (LSD1), is a floral activator that downregulates methylation of H3K4 in FLC chromatin and represses FLC expression.Citation18,Citation19 Interestingly, bacteria expressing recombinant FLD protein did not demethylate H3K4me2, inferring that the demethylase activity requires additional co-factors as are necessary for LSD1.Citation18,Citation20 Together, these results suggest that SIZ1-mediated SUMO modification of FLD may affect interactions between FLD and co-factors, which is necessary for FLC chromatin modification.
Despite our results that implicate SA in flowering time control, how SIZ1 regulates SA accumulation and the identity of the effectors involved remain to be discovered. In addition, it remains to be determined if SIZ1 is involved in other mechanisms that modulate FLD activity and FLC expression, or the function of other autonomous pathway determinants.
Figures and Tables
Figure 1 Model of how SUMO conjugation and deconjugation regulate plant development in Arabidopsis. SIZ1 and Avr proteins regulate biosynthesis and accumulation of SA, a plant stress hormone that is involved in plant innate immunity, leaf development and regulation of flowering time. SA promotes transition to flowering may through AGL15/AGL18 dependent and independent pathways. FLC expression is activated by FRIGIDA but repressed by the autonomous pathway gene FLD, and SIZ1-mediated sumoylation of FLD represses its activity. Lines with arrows indicate upregulation (activation), and those with bars identify downregulation (repression).
Addendum to:
References
- Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, Back D, Kim DH, Jeoung JC, et al. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 2007; 49:79 - 90
- Lois LM, Lima CD, Chua NH. Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell 2003; 15:1347 - 1359
- Murtas G, Reeves PH, Fu YF, Bancroft I, Dean C, Coupland G. A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of small ubiquitin-related modifier conjugates. Plant Cell 2003; 15:2308 - 2319
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, Yun DJ, Hasegawa PM. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 2005; 102:7760 - 7765
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM. SIZ1-Mediated Sumoylation of ICE1 Controls CBF3/DREB1A Expression and Freezing Tolerance in Arabidopsis. Plant Cell 2007; 19:1403 - 1414
- Miura K, Jin JB, Hasegawa PM. Sumoylation, a post-translational regulatory process in plants. Curr Opin Plant Biol 2007; 10:495 - 502
- Yoo CY, Miura K, Jin JB, Lee J, Park HC, Salt DE, Yun DJ, Bressan RA, Hasegawa PM. SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol 2006; 142:1548 - 1558
- Catala R, Ouyang J, Abreu IA, Hu Y, Seo H, Zhang X, Chua NH. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 2007; 19:2952 - 2966
- Jin JB, Jin YH, Lee J, Miura K, Yoo CY, Kim WY, Van OM, Hyun Y, Somers DE, Lee I, Yun DJ, Bressan RA, Hasegawa PM. The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J 2008; 53:530 - 540
- Balasubramanian S, Sureshkumar S, Lempe J, Weigel D. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet 2006; 2:106
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA 2000; 97:11655 - 11660
- Adamczyk BJ, Lehti-Shiu MD, Fernandez DE. The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J 2007; 50:1007 - 1019
- Orth K, Xu Z, Mudgett MB, Bao ZQ, Palmer LE, Bliska JB, Mangel WF, Staskawicz B, Dixon JE. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 2000; 290:1594 - 1597
- Hotson A, Chosed R, Shu H, Orth K, Mudgett MB. Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol Microbiol 2003; 50:377 - 389
- Roden J, Eardley L, Hotson A, Cao Y, Mudgett MB. Characterization of the Xanthomonas AvrXv4 effector, a SUMO protease translocated into plant cells. Mol Plant Microbe Interact 2004; 17:633 - 643
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA 2003; 100:8024 - 8029
- Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol 2004; 42:185 - 209
- Jiang D, Yang W, He Y, Amasino RM. Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 2007; 19:2975 - 2987
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004; 119:941 - 953
- Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 2005; 437:432 - 435