Abstract
Plastids rely on multiple phosphate (Pi) transport activities to support and control a wide range of metabolic processes, including photosynthesis and carbon partitioning. Five of the six members of the PHT4 family of Pi transporters in Arabidopsis thaliana (PHT4;1-PHT4;5) are confirmed or predicted plastid proteins. As a step towards identifying the roles of individual PHT4 Pi transporters in chloroplast and non-photosynthetic plastid Pi dynamics, we used promoter-reporter gene fusions and quantitative RT-PCR studies, respectively, to determine spatial and diurnal gene expression patterns. PHT4;1 and PHT4;4 were both expressed predominantly in photosynthetic tissues, although expression of PHT4;1 was circadian and PHT4;4 was induced by light. PHT4;3 and PHT4;5 were expressed mainly in leaf phloem. PHT4;2 was expressed throughout the root, and exhibited a diurnal pattern with peak transcript levels in the dark. The remaining member of this gene family, PHT4;6, encodes a Golgi-localized protein and was expressed ubiquitously. The overlapping but distinct expression patterns for these genes suggest specialized roles for the encoded transporters in multiple types of differentiated plastids. Phylogenetic analysis revealed conservation of each of the orthologous members of the PHT4 family in Arabidopsis and rice, which is consistent with specialization, and suggests that the individual members of this transporter family diverged prior to the divergence of monocots and dicots.
Introduction
Dynamic control of stromal inorganic phosphate (Pi) levels is central to the specialized metabolic functions of differentiated plastids. Notably, the concentration of Pi in the chloroplast stroma is tightly coordinated with environmental conditions to modulate both photosynthesis and the subsequent partitioning of fixed carbon.Citation1,Citation2 Pi concentrations in amyloplasts also are held within a critical limit to prevent inhibition of starch biosynthesis.Citation3 For each plastid type, Pi concentrations are controlled through a combination of metabolic recycling in the stroma and surrounding cytosol, and the transport of Pi across the plastid limiting membrane. Recent data suggest that similar processes also link the Pi status of the chloroplast stroma and thylakoid lumen.Citation4
Plastidic Pi transport is generally attributed to members of the plastidic phosphate translocator (pPT) family.Citation5 These proteins are located in the inner envelope membrane and mediate strict counter-exchange of Pi for phosphorylated C3, C5 or C6 compounds. The triose phosphate/Pi translocator (TPT) was the first pPT protein to be identified, and it is expressed almost exclusively in photosynthetic tissues where it catalyzes transport of cytosolic Pi into the chloroplast in exchange for triose phosphates, the end products of photosynthesis.Citation6 This activity represents the major pathway for carbon allocation to the cytosol during the day as well as the primary route for Pi import into the chloroplast. Related members of the pPT family include the phosphoenolpyruvate/Pi translocator (PPT), glucose 6-phosphate/Pi translocator (GPT) and xylulose 5-phosphate/Pi translocator (XPT).Citation7–Citation9 In contrast to TPT, these translocators export Pi from plastids in exchange for cytosolic metabolites that serve as precursors for biosynthetic processes within the stroma. Moreover, PPT and XPT are expressed in both photosynthetic and heterotrophic tissues, and GPT expression is restricted to heterotrophic tissues.Citation10
Data mining of plant genome sequences coupled with plastid envelope proteomics has revealed additional classes of plastidic Pi transporters that are unrelated to members of the pPT family. PHT2;1 was identified in Arabidopsis based on its similarity with Na+/Pi symporters from mammals and fungi.Citation11 Functional analyses in yeast, however, suggest that PHT2 proteins catalyze H+-dependent Pi transport.Citation11–Citation13 GFP translational fusions with PHT2 proteins from Arabidopsis, Medicago truncatula, spinach and potato are targeted to the chloroplast envelope,Citation12–Citation15 and localization within the inner envelope membrane is supported by subcellular proteomics and membrane fractionation/immunodetection.Citation14 In addition to a putative role in Pi import into the chloroplast, the presence of PHT2;1 transcripts within the root stele suggests that the encoded protein also functions in a subset of non-photosynthetic plastids.Citation15
The Arabidopsis genome also encodes six PHT4 proteins, all of which mediate Pi transport in yeast with high specificity.Citation16 The effects of pH and protonophores on transport activities suggest that PHT4 transporters, like PHT2 proteins, catalyze H+-dependent Pi transport. Functional studies in Escherichia coli suggest that at least one of these, PHT4;1, can also mediate Na+-dependent Pi transport.Citation4 Bioinformatics and localization of PHT4-GFP protein fusions indicate that five members of this family (PHT4;1 through PHT4;5) are targeted to plastids,Citation16–Citation18 and the sixth, PHT4;6, resides in the Golgi apparatus.Citation16 Proteomic analysis of the chloroplast envelope and envelope membrane fractionation studies confirmed that PHT4;4 is located in the chloroplast inner envelope membrane.Citation4,Citation17,Citation18 In contrast, PHT4;1 is located in the thylakoid membrane.Citation4 Despite the localization of these two proteins to chloroplast membranes, transcripts for all of the Arabidopsis PHT4 genes have been detected in roots as well as leaves suggesting that the encoded plastid-targeted proteins also function in non-photosynthetic plastids.Citation16 Previous studies have not distinguished whether these transporters are redundant or have unique specificities for different plastid types. Here we present evidence for differences in spatial expression patterns and regulation with respect to light and circadian rhythm, which suggest specialized roles for the encoded Pi transporters in different plastid types and highlight the diversity of plastids that are present in heterotrophic tissues. Moreover, comparative analyses of Arabidopsis and rice PHT4 protein sequences suggest that these transporters and their respective roles are conserved.
Results
Tissue specificity of Arabidopsis PHT4 genes.
To analyze the spatial expression patterns of the PHT4 genes throughout plant development we used a promoter-reporter strategy in which each promoter was cloned upstream of the β-glucuronidase (GUS) reporter gene. The transcriptional fusion constructs were introduced into Arabidopsis plants, and progeny of the transgenic plants were evaluated for GUS activity using a histochemical assay.
PHT4;1 and PHT4;4 promoters conferred nearly identical GUS expression patterns. Reporter activity was detected throughout the green tissues of seedlings (), and in sepals of mature flowers (). In sections prepared from rosette leaves, GUS activity appeared to be present in all cell types but was not uniformly distributed in the epidermis (). To examine expression in epidermal cells more directly, we isolated and stained leaf epidermal peels. GUS expression was detected only in guard and subsidiary cells (). In all cases, GUS activity co-localized with chlorophyll autofluorescence (red signal, ), indicating that PHT4;1 and PHT4;4 are expressed predominantly in chloroplasts of photosynthetic cells. GUS activity was, however, also detected at low levels in the root stele of seedlings containing the PHT4;1 promoter-GUS fusion (). Despite previous detection of PHT4;4 transcripts in roots by RT-PCR,Citation16 no GUS activity was detected in roots of plants that carried the PHT4;4 promoter-GUS fusion.
GUS expression driven by the PHT4;2 promoter was detected throughout the root (), but was not detected in either leaf or floral tissues.
Plants harboring the PHT4;3 and PHT4;5 promoter-GUS fusions also exhibited similar expression patterns, but these patterns clearly differed from those directed by the PHT4;1 and PHT4;4 promoters. In leaves and cotyledons, GUS expression driven by the PHT4;3 and PHT4;5 promoters was restricted to the veins (). Differential interference contrast (DIC) imaging of transverse leaf sections revealed that expression in these organs was limited to the phloem portion of the vascular bundle (). Although tissue specificity for the PHT4;3 and PHT4;5 promoters appeared identical in leaves, expression differed in roots and flowers. In roots, weak GUS activity was detected at the root tip in plants carrying the PHT4;3 promoter-GUS construct (), whereas no activity was detected in roots harboring the PHT4;5 reporter construct. In flowers, the PHT4;3 promoter generated no detectable staining, but the PHT4;5 promoter directed GUS expression that was restricted to sepals, essentially the same pattern seen with the PHT4;1 and PHT4;4 promoters ().
PHT4;6 is the only member of the PHT4 family that is targeted to the Golgi apparatus rather than plastids.Citation16 We examined the tissue specificity of this promoter to determine whether PHT4;6 might serve as a marker for potential heterogeneity of Golgi functions in different plant tissues. No such specificity was observed. GUS activity was detected in every part of transgenic seedlings (), as well as sepals, stamens and carpels of mature flowers ().
Light and circadian control of PHT4 gene expression.
TPT and PHT2;1 are expressed primarily in photosynthetic tissues, and expression is induced by light.Citation12,Citation15,Citation24 To determine whether light also influences expression of PHT4 genes, we used quantitative RT-PCR to monitor transcript levels in rosette leaves of 3-wk-old plants that had been held in the dark for 3 d and at defined time points after re-exposure to light (). PHT2;1 served as a positive control for light-induced expression.Citation12,Citation15 As expected, PHT2;1 transcript levels increased rapidly after exposure to light. PHT4;1 and PHT4;4 transcript levels also increased during the 7 hr light treatment, 20-fold and 10-fold, respectively, although transcripts for both genes accumulated only after a 3 hr lag. In contrast, the light treatment had no obvious effect on expression of PHT4;3, PHT4;5 or PHT4;6. PHT4;2 transcript levels were extremely low, i.e., maximum levels were less than 2% of the lowest value obtained for the other genes (data not shown). This low expression was consistent with promoter-GUS results indicating that PHT4;2 is expressed almost exclusively in roots (). Consequently, we evaluated PHT4;2 transcript levels in roots harvested from the same plants. As shown in , PHT4;2 transcript levels decreased nearly 80% during the light treatment, and like PHT4;1 and PHT4;4, the change in transcript abundance occurred only after a 3 hr lag.
Although PHT2;1 expression is not under circadian control,Citation15 it was possible that the changes in PHT4;1, PHT4;4 and PHT4;2 transcript levels during the light treatment () reflect rhythmic expression rather than a response to light. To distinguish between these possibilities, transcript levels in rosette leaves of 3-wk-old plants maintained with a 14 hr light: 10 hr dark cycle (LD) were compared to those in plants that had been transferred to constant light (LL). Plants were harvested at time points corresponding to the midpoint and end of the subjective light and dark phases of two consecutive diurnal cycles. As shown in , PHT4;1 and PHT4;4 both exhibited a diurnal expression pattern under LD conditions, but this rhythmic pattern persisted in LL only for PHT4;1. Thus, PHT4;1 exhibits a circadian expression pattern with peak expression during the light phase of the diurnal cycle. Consistent with this circadian pattern, two copies of the CIRCADIAN CLOCK ASSOCIATED 1-binding site (CBS: AAAAATCT), which is important for morning-specific circadian expression,Citation25,Citation26 are located in the PHT4;1 promoter. One CBS is located −17 bp relative to the translation start, and the other is located −281 bp and is on the complementary strand. No CBS motifs were found in the promoters of the other PHT4 genes.
In contrast to the circadian expression of PHT4;1, expression of PHT4;4 appears to be induced by light, both after a long dark period () and in standard LD conditions (). PHT4;3 and PHT4;5 transcript levels, which were unaffected by light exposure treatment after a long dark treatment (), varied with the light/dark cycle in LD but not under LL conditions (). Transcripts for all three of these genes were more abundant in the light than dark.
To determine whether PHT4;2 expression also exhibited a diurnal pattern, we compared transcript levels in roots of plants grown under LD and LL conditions. As shown in , transcript levels appeared rhythmic in LD with a 2.5-fold difference between the light and dark. However, the phase of this rhythm was opposite to those of the other genes. That is, PHT4;2 transcripts were more abundant in the dark than light. This expression pattern did not persist in LL suggesting that the pattern in LD is related to exposure of the shoot tissues to light rather than circadian control.
Conservation of the PHT4 family in Arabidopsis and rice.
Searches of the GenBank sequence database revealed putative PHT4 homologs in multiple plant species, including rice, tomato, maize, grape and barley, suggesting that these transporters are widely conserved. However, it is difficult to determine orthology because this gene family may have undergone differential expansion in each species. The availability of the complete genome sequences for Arabidopsis and rice allowed us to address this issue for two distantly related plant species. We identified seven protein sequences through searches of the TIGR rice genome database (http://rice.tigr.org/)Citation27 that shared 70–80% similarity to Arabidopsis PHT4 proteins. Iterative searches with the rice sequences revealed no additional homologs, indicating that these seven proteins comprise the rice PHT4 family. To estimate the phylogenetic relationship between the Arabidopsis and rice proteins, the sequences were aligned with ClustalX and M-Coffee,Citation28 and neighbor joining was used to generate an unrooted phylogram (). The topology of this phylogram is supported by 100% bootstrap values. Furthermore, maximum parsimony analysis with the same alignments yielded trees with identical topologies (not shown). Six groups of proteins are inferred from the phylogram, each consisting of a single Arabidopsis protein and, in all but one group, a single orthologous rice protein. The exception was the group containing PHT4;6, which includes two paralogous rice sequences. The presence of both Arabidopsis and rice proteins in each group indicates that divergence of the PHT4 family occurred prior to the divergence of monocots and dicots.
Like the Arabidopsis PHT4 proteins, all of the rice orthologs have poorly conserved, N-terminal sequences that share features with organellar targeting sequences. Predictions of the subcellular localization of these proteins using TargetP,Citation29 as well as consensus predictions derived from multiple other programs (ARAMEMNON database, http:arememnon.botanik.uni-koeln.de/), suggest that each rice protein is targeted to the same organelle as its corresponding Arabidopsis ortholog (). The locus identifiers for the rice PHT4 sequences and the corresponding assignments from the International Rice Genome Sequencing Project (IRGSP, http://rgp.dna.affrc.go.jp/IRGSP/) are also listed in .
Discussion
Phylogenetic analysis suggests that members of the PHT4 Pi transporter family diverged prior to the monocot-dicot split that is estimated to have occurred 140–150 Myr ago.Citation30 Surprisingly, all six of the ancestral members have been preserved in Arabidopsis and rice, and only one ortholog appears to have undergone duplication. This level of conservation was unexpected given that multiple, large-scale duplication and deletion events have occurred since the divergence of monocots and dicots.Citation31,Citation32 Indeed, a similar analysis of PHT1 Pi transporters revealed orphan homologs in both Arabidopsis and rice, as well as differential expansion of ancestral orthologs.Citation33 The simplest explanation for the conservation of individual PHT4 orthologs is that each has unique physiological roles. The overlapping but distinct expression patterns for the Arabidopsis genes support this hypothesis.
The PHT4;1 promoter was active predominantly in photosynthetic tissues, and in the leaf epidermis, activity was restricted to cells that contain chlorophyll. This tissue and cell specificity is consistent with the recent finding that the protein is targeted to the chloroplast thylakoid membrane.Citation4 This location positions PHT4;1 for a role in recycling Pi from the thylakoid lumen to the stroma. Pi is generated in the thylakoid lumen through hydrolysis of nucleotides, including the PsbO-mediated hydrolysis of GTP, which regulates dephosphorylation and turnover of the photosystem II reaction center D1 protein.Citation34,Citation35 Coordination of PHT4;1 activity with D1 protein turnover is an intriguing possibility given that the prerequisite events, expression of PHT4;1 and phosphorylation of the D1 protein, are both controlled by a circadian rhythm with peak activities in the light.Citation36 Circadian regulation is critical for many aspects of photosynthesis and plant growth,Citation37 but to our knowledge, this is the first example of circadian control of a plant Pi transporter gene.
The stimulatory effect of low pH on Pi transport catalyzed by PHT4;1 in heterologous systems would favor export of Pi from the acidic lumen.Citation4,Citation16 However, at this time we cannot rule out the possibility that PHT4;1 transports stromal Pi into the lumen. It has been suggested that if such an activity sequesters stromal Pi below the Km of the thylakoid ATP synthase, the resulting decrease in proton conductivity could serve as a means of down regulating photosynthetic light capture.Citation4,Citation38
The PHT4;1 promoter also exhibited weak activity in the root stele (), which corroborates previous results indicating that endogenous transcripts are present in roots at low levels.Citation16 The physiological significance of this expression is unclear because root plastids have limited internal membranes,Citation39 and the metabolic contributions of plastids within the root stele have not been defined. We hypothesize that the internal membranes of these plastids represent a metabolically active compartment and that PHT4;1 is involved in its Pi homeostasis.
Chloroplasts rely on the import of Pi from the cytosol to support the synthesis of ATP through photophosphorylation and to control the subsequent partitioning of fixed carbon. Pi import is mediated mainly by TPT with strict counterexchange of stromal triose phosphates.Citation40 Surprisingly, plants with reduced levels of TPT have no substantial growth phenotype or reduction in photosynthetic capacity when grown under ambient conditions.Citation41–Citation46 Such plants do, however, exhibit increased rates of starch turnover and export of neutral sugars to compensate for the defect in carbon allocation, which suggests that redundant or compensatory mechanisms also exist for the coupled defect in Pi import. PHT4;4 and PHT2;1 are candidates for this activity. Like TPT, both of these Pi transporters are located in the chloroplast inner envelope membrane.Citation14,Citation18 Also, transcript levels for the genes increased when plants were exposed to light, although the delay in PHT4;4 transcript accumulation may reflect regulation by photosynthates rather than a direct response to light. It is formally possible that PHT4;4 and PHT2;1 also maintain stromal Pi concentrations through export. That is, these transporters may serve as two-way valves with transport direction dependent on the Pi electrochemical gradient and proton-motive force. Such flexibility would enable fine control of stromal Pi levels, which may be necessary to sustain high rates of transitory starch synthesis. Experimental evaluation of this hypothesis is needed.
Unlike chloroplasts, non-photosynthetic plastids cannot synthesize ATP or precursor metabolites for anabolic processes such as the synthesis of starch, fatty acids and amino acids.Citation47 The import and subsequent assimilation of these compounds can lead to an imbalance in stromal Pi, which must be countered by a Pi export activity that is not directly coupled to the transport of phosphorylated carbon compounds, i.e., unidirectional Pi transport.Citation48 Several of the PHT4 transporters, as well as PHT2;1, may contribute to this role in different heterotrophic tissues. In roots, GUS activity driven by the PHT4;2 promoter was observed throughout the entire organ, whereas expression under the control of the PHT4;3 and PHT2;1 promoters was restricted to the root cap and stele, respectively. These expression patterns suggest that PHT4;2 functions in all root plastids and that Pi transport catalyzed by PHT4;3 and PHT2;1 may be needed to supplement that of PHT4;2 in distinct plastid types, including amyloplasts in root cap columella cells. Although PHT4;4 and PHT4;5 transcripts have been detected in roots by RT-PCRCitation16 and the corresponding promoter-GUS lines described here exhibited strong activity in green tissues, no GUS activity was detected in roots. This suggests that PHT4;4 and PHT4;5 are expressed in root tissues at levels below the limit of detection by histochemical staining, and therefore, may have limited roles in root plastids.
Non-photosynthetic plastids are not typically associated with aerial tissues. However, such plastids have been identified in the sieve elements and companion cells that comprise phloem.Citation49,Citation50 Promoter-GUS fusions used in this study revealed that PHT4;3 and PHT4;5 were expressed in the phloem tissue of leaves and cotyledons, but not roots. Expression of both PHT4;3 and PHT4;5 in leaves was diurnal with transcripts more abundant in the light. Transcript abundance was not induced by light after a 3 d dark treatment suggesting that the diurnal pattern may be related to instability of the transcripts in the dark. Little has been reported on the metabolic functions of phloem plastids, although they may be the primary sites of tryptophan biosynthesis,Citation51 and at least some of these plastids accumulate starch.Citation50 Given that amino acid and starch synthesis are ATP consuming and Pi liberating processes, we hypothesize that PHT4;3 and PHT4;5 contribute to Pi export from phloem plastids.
In summary, the results of this study have revealed differences in the spatial expression patterns and regulation of the plastid-localized members of the Arabidopsis PHT4 Pi transporter family. These differences provide some insight into the potential roles of these proteins in chloroplasts and in an unexpectedly wide range of non-photosynthetic plastids in root and phloem tissues. The remarkable similarity of the PHT4 family structure between Arabidopsis and rice suggests that the roles for the individual family members have been conserved since the divergence of monocots and dicots.
Materials and Methods
Plant growth conditions.
Arabidopsis thaliana (L.) Heynh. (ecotype Col-0) plants were grown in chambers at 21°C with 70% relative humidity and, unless specified otherwise, a 14 h light (150 µmol m−2 s−1): 10 h dark photocycle. Plants were grown in soil for general propagation. For gene expression studies, plants were grown hydroponically or on agar-solidified, half-strength MS mediumCitation19 as described previously.Citation16
Construction and analysis of PHT4 promoter-GUS fusions.
Promoters for each PHT4 gene were amplified from Arabidopsis genomic DNA using Pfx high-fidelity polymerase (Invitrogen, Carlsbad, CA) then cloned in the binary vector pBI101.Citation1 upstream of the β-glucuronidase (GUS) gene to generate transcriptional fusions. Promoter sequences correspond to the region immediately upstream of the respective ATG start codons: 1000 bp for PHT4;1, PHT4;2, PHT4;3 and PHT4;4, 623 bp for PHT4;5, and 1682 bp for the PHT4;6. The PHT4;5 promoter extends to the stop codon of the neighboring gene (At5g20390).
Promoter-GUS constructs were introduced into Agrobacterium tumefaciens strain GV3101, and the resulting strains were used to transform Arabidopsis via the floral dip procedure.Citation20 Transgenic seedlings were selected on half-strength MS medium containing 25 µg ml−1 kanamycin. At least 12 independent lines for each construct were examined for GUS activity by histochemical detection.Citation21,Citation22 Briefly, seedlings were vacuum infiltrated with assay buffer (50 mM sodium phosphate pH 7.0, 0.1% (v/v) Triton X-100, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, 10 mM EDTA) containing 0.05% (w/v) 5-bromo-4-chloro-3-indolyl glucuronide (X-gluc) then incubated in the dark at 37°C overnight. Green tissues were destained with 70% ethanol prior to observation. To prepare sections, tissues were briefly fixed in 4% formaldehyde prior to staining. After staining, tissues were fixed in assay buffer containing 4% formaldehyde and 0.5% glutaraldehyde then dehydrated in an ethanol series and embedded in Steedman's wax (polyethylene glycol 400 distearate/ 1-hexadecanol, [9:1, w/w]).Citation23 Embedded tissue was cut into 10 µm sections using a rotary microtome.
Quantitative RT-PCR analysis.
Total RNA was isolated from rosette leaves or roots of 3-wk-old plants with TRI reagent (Sigma-Aldrich, St. Louis, MO), and traces of DNA were removed with TURBO DNA-free (Ambion, Austin, TX). Two biological replicates consisting of tissues pooled from two plants were used for each analysis. One microgram RNA was used to make cDNA with SuperScript first-strand cDNA synthesis kit (Invitrogen, Carlsbad, CA). PCR was performed with Power SYBR Green Master Mix (Applied Biosystems, Foster City, CA) using the ABI Prism 7500 sequence detection system (Applied Biosystems). Expression levels were normalized to EIF-4A2 (At1g54270) and fold changes were calculated using the ΔΔCT method. Primers used for real-time PCR were described previously.Citation16
Abbreviations
| Pi | = | inorganic phosphate |
| LL | = | continuous light |
| LD | = | light-dark cycle |
Figures and Tables
Figure 1 Localization of promoter-GUS gene fusions in transgenic A. thaliana plants. (a) Seedling representative of PHT4;1-GUS and PHT4;4-GUS plants. (b) PHT4;2-GUS seedling. (c) Flower representative of PHT4;1-GUS, PHT4;4-GUS and PHT4;5-GUS plants. (d) PHT4;6-GUS flower. (e) Leaf and cotyledon representative of PHT4;3-GUS and PHT4;5-GUS plants showing vascular-specific GUS activity. (f) PHT4;6-GUS seedling. (g) Root of PHT4;1-GUS seedling. (h) Root of PHT4;3-GUS seedling. (i) Transverse leaf section representative of PHT4;1-GUS and PHT4;4-GUS plants. (j) Leaf epidermal peel representative of PHT4;1-GUS and PHT4;4-GUS plants. (k) Chlorophyll autofluorescence in epidermal peel shown in (j). (l) Transverse leaf section representative of PHT4;3-GUS and PHT4;5-GUS plants showing GUS activity in phloem. Bars in (g, h) = 100 µm; bars in (i-l) = 20 µm.
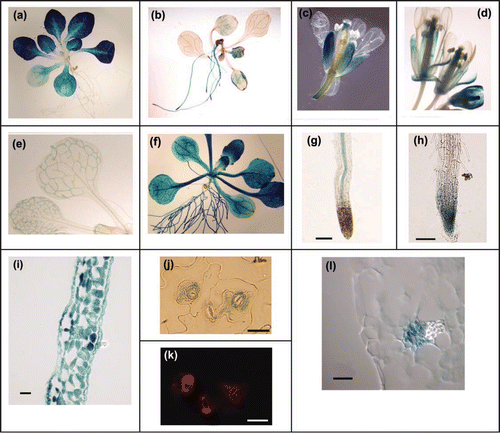
Figure 2 Effect of light on expression of PHT4 genes. Mature plants were held in the dark for 3 d before re-exposure to light. Expression levels were determined at the indicated time points by real-time RT-PCR and normalized to EIF-4A2. For each gene, expression values are relative to the maximum value, which was set at 1. The values plotted are averages of two biological replicates, and error bars indicate the replicate values. For analysis of PHT4;2, RNA was isolated from root tissues. For all other genes, RNA was isolated from rosette leaves. PHT2;1 serves as a positive control for light induction.
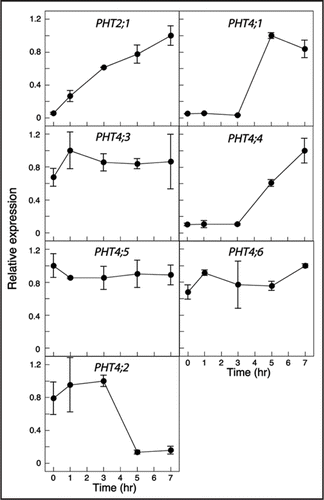
Figure 3 Expression of PHT4 genes under light/dark and constant light conditions. Expression levels were measured at the indicated times by real-time RT-PCR and normalized to EIF-4A2. For each gene, expression values are relative to the maximum value, which was set at 1. The values plotted are averages of two biological replicates, and error bars indicate the replicate values. The light and dark bars at the top of the figure indicate the respective light conditions. For analysis of PHT4;2, RNA was isolated from root tissues. For all other genes, RNA was isolated from rosette leaves.
Figure 4 Neighbor joining tree of Arabidopsis and rice PHT4 Pi transporter proteins. Rice sequences are indicated by TIGR locus assignments. The numbers at the branches of the tree are bootstrap values (1,000 replicates). Scale bar indicates substitutions per site.

Table 1 Members of the rice PHT4 family
Acknowledgements
We thank Catherine Wussler for her participation in constructing promoter-GUS fusion clones; Sara Almendarez, Heather Belveal, Sara Fradenburgh and Diwen Chen for assistance with plant care; and Tom McKnight, Pallavi Mukherjee and Cornelia Spetea Wiklund for critical reading of this manuscript. This work was supported by a grant from the National Science Foundation (IOS-0416443).
References
- Sharkey TD. Photosynthesis in intact leaves of C3 plants: physics, physiology and rate limitations. Bot Revwalk 1985; 51:53 - 105
- Walker DA, Sivak MN. Photosynthesis and phosphate: a cellular affair?. Trends Biochem Sci 1986; 11:176 - 179
- Preiss J. Regulation of the biosynthesis and degradation of starch. Annu Rev Plant Physiol Plant Mol Biol 1982; 33:431 - 454
- RuizPavón LR, Lundh F, Lundin B, Mishra A, Persson BL, Spetea C. Arabidopsis ANTR1 is a thylakoid Na+-dependent phosphate transporter: functional characterization in Escherichia coli. J Biol Chem 2008; 283:13520 - 13527
- Knappe S, Flügge UI, Fischer K. Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol 2003; 131:1178 - 1190
- Flügge UI, Fischer K, Gross A, Sebald W, Lottspeich F, Eckerskorn C. The triose phosphate-3-phosphoglycerate-phosphate translocator from spinach chloroplasts: nucleotide sequence of a full-length cDNA clone and import of the in vitro synthesized precursor protein into chloroplasts. EMBO J 1989; 8:39 - 46
- Eicks M, Maurino V, Knappe S, Flügge UI, Fischer K. The plastidic pentose phosphate translocator represents a link between the cytosolic and the plastidic pentose phosphate pathways in plants. Plant Physiol 2002; 128:512 - 522
- Fischer K, Kammerer B, Gutensohn M, Arbinger B, Weber A, Häusler RE, et al. A new class of plastidic phosphate translocators: a putative link between primary and secondary metabolism by the phosphoenolpyruvate/phosphate antiporter. Plant Cell 1997; 9:453 - 462
- Kammerer B, Fischer K, Hilpert B, Schubert S, Gutensohn M, Weber A, et al. Molecular characterization of a carbon transporter in plastids from heterotrophic tissues: the glucose 6-phosphate/phosphate antiporter. Plant Cell 1998; 10:105 - 117
- Flügge UI. Functional genomics of phosphate antiport systems in plastids. Physiol Plantarum 2003; 118:475 - 482
- Daram P, Brunner S, Rausch C, Steiner C, Amrhein N, Bucher M. Pht2;1 encodes a low-affinity phosphate transporter from Arabidopsis. Plant Cell 1999; 11:2153 - 2166
- Versaw WK, Harrison MJ. A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 2002; 14:1751 - 1766
- Zhao L, Versaw WK, Liu J, Harrison MJ. A phosphate transporter from Medicago truncatula is expressed in the photosynthetic tissues of the plant and is located in the chloroplast envelope. New Phytol 2003; 157:291 - 302
- Ferro M, Salvi D, Riviere-Rolland H, Vermat T, Seigneurin-Berny D, Grunwald D, et al. Integral membrane proteins of the chloroplast envelope: identification and subcellular localization of new transporters. Proc Natl Acad Sci USA 2002; 99:11487 - 11492
- Rausch C, Zimmermann P, Amrhein N, Bucher M. Expression analysis suggests novel roles for the plastidic phosphate transporter Pht2;1 in auto- and heterotrophic tissues in potato and Arabidopsis. Plant J 2004; 39:13 - 28
- Guo B, Jin Y, Wussler C, Blacaflor EB, Motes CM, Versaw WK. Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol 2008; 177:889 - 898
- Ferro M, Salvi D, Brugiere S, Miras S, Kowalski S, Louwagie M, et al. Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol Cell Proteomics 2003; 2:325 - 345
- Roth C, Menzel G, Petetot JM, Rochat-Hacker S, Poirier Y. Characterization of a protein of the plastid inner envelope having homology to animal inorganic phosphate, chloride and organic-anion transporters. Planta 2004; 218:406 - 416
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plantarum 1962; 15:473 - 497
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 1998; 16:735 - 743
- Jefferson RA, Kavanaugh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versitile gene fusion marker in higher plants. EMBO J 1987; 6:3901 - 3907
- Vitha S, Benes K, Phillips JP, Gartland KMA. Gartland KMA, Davey MR. Histochemical GUS analysis. Agrobacterium Protocols 1995; Totowa, NJ, USA Humana Press 185 - 193
- Steedman HF. Polyester wax; a new ribboning embedding medium for histology. Nature 1957; 179:1345
- Schulz B, Frommer WB, Flügge UI, Hummel S, Fischer K, Willmitzer L. Expression of the triose phosphate translocator gene from potato is light dependent and restricted to green tissues. Mol Gen Genet 1993; 238:357 - 361
- Michael TP, McClung CR. Phase-specific circadian clock regulatory elements in Arabidopsis. Plant Physiol 2002; 130:627 - 638
- Wang Z-Y, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM. A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 1997; 9:491 - 507
- Yuan Q, Ouyang S, Wang A, Zhu W, Maiti R, Lin H, et al. The institute for genomic research Osa1 rice genome annotation database. Plant Physiol 2005; 138:18 - 26
- Moretti S, Armougom F, Wallace IM, Higgins DG, Jongeneel CV, Notredame C. The M-Coffee web server: a meta-method for computing multiple sequence alignments by combining alternative alignment methods. Nucleic Acids Res 2007; 35:W645 - W648
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protocols 2007; 2:953 971
- Chaw SM, Chang CC, Chen HL, Li WH. Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol 2004; 58:424 - 441
- Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA 2004; 101:9903 - 9908
- Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y. The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA 2002; 99:13627 - 13632
- Paszkowski U, Kroken S, Roux C, Briggs SP. Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 2002; 99:13324 - 13329
- Lundin B, Hansson M, Schoefs B, Vener AV, Spetea C. The Arabidopsis PsbO2 protein regulates dephosphorylation and turnover of the photosystem II reaction centre D1 protein. Plant J 2007; 49:528 - 539
- Lundin B, Thuswaldner S, Shutova T, Eshaghi S, Samuelsson G, Barber J, et al. Subsequent events to GTP binding by the plant PsbO protein: structural changes, GTP hydrolysis and dissociation from the photosystem II complex. Bioch Biophys Acta 2007; 1767:500 - 508
- Booij-James IS, Swegle WM, Edelman M, Mattoo AK. Phosphorylation of the D1 photosystem II reaction center protein is controlled by an endogenous circadian rhythm. Plant Physiol 2002; 130:2069 - 2075
- Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 2005; 309:630 - 633
- Takizawa K, Kanazawa A, Kramer DM. Depletion of stromal Pi induces high ‘energy-dependent’ antenna exciton quenching (qE) by decreasing proton conductivity at CFo-CF1 ATP synthase. Plant Cell Environ 2008; 31:235 - 243
- Usami T, Mochizuki N, Kondo M, Nishimura M, Nagatani A. Cryptochromes and phytochromes synergistically regulate Arabidopsis root greening under blue light. Plant Cell Physiol 2004; 45:1798 - 1808
- Fliege R, Flügge UI, Werdan K, Heldt HW. Specific transport of inorganic phosphate, 3-phosphoglycerate and triosephosphates across the inner membrane of the envelope in spinach chloroplasts. Bioch Biophys Acta 1978; 502:232 - 247
- Barnes SA, Knight JS, Gray JC. Alteration of the amount of the chloroplast phosphate translocator in transgenic tobacco affects the distribution of assimilate between starch and sugar. Plant Physiol 1994; 106:1123 - 1129
- Häusler RE, Schlieben NH, Schultz B, Flügge U. Compensation of the decreased triose phosphate/phosphate translocator activity by accelerated starch turnover and glucose transport in transgenic tobacco. Planta 1998; 204:366 - 376
- Heineke D, Kruse A, Flügge U, Frommer WB, Riesmeier JW, Willmitzer L, et al. Effect of antisense repression of the chloroplast triose-phosphate translocator on photosynthetic metabolism in transgenic potato plants. Planta 1994; 193:174 - 180
- Riesmeier JW, Flügge UI, Schulz B, Heineke D, Heldt HW, Willmitzer L, et al. Antisense repression of the chloroplast triose phosphate translocator affects carbon partitioning in transgenic potato plants. Proc Natl Acad Sci USA 1993; 90:6160 - 6164
- Schneider A, Häusler RE, Kolukisaoglu Ü, Kunze R, van der Graaff E, Schwacke R, et al. An Arabidopsis thaliana knock-out mutant of the chloroplast triose phosphate/phosphate translocator is severely compromised only when starch synthesis, but not starch mobilisation is abolished. Plant J 2002; 32:685 - 699
- Walters RG, Ibrahim DG, Horton P, Kruger NJ. A mutant of Arabidopsis lacking the triosephosphate/phosphate translocator reveals metabolic regulation of starch breakdown in the light. Plant Physiol 2004; 135:891 - 906
- Neuhaus HE, Emes MJ. Nonphotosynthetic metabolism in plastids. Annu Rev Plant Physiol Plant Mol Biol 2000; 51:111 - 140
- Neuhaus HE, Maass U. Unidirectional transport of orthophosphate across the envelope of isolated cauliflower-bud amyloplasts. Planta 1996; 198:542 - 548
- Behnke HD. Plastids in sieve elements and their companion cells. Planta 1973; 110:321 - 328
- Behnke HD. Distribution and evolution of forms and types of sieve-element plastids in the dicotyledons. Aliso 1991; 3:167 - 182
- Lu H, McKnight TD. Tissue-specific expression of the beta-subunit of tryptophan synthase in Camptotheca acuminata, an indole alkaloid-producing plant. Plant Physiol 1999; 120:43 - 52