Abstract
In animal cells, nitric oxide and NO-derived molecules have been shown to mediate post-translational modifications such as S-nitrosylation and protein tyrosine nitration which are associated with cell signalling and pathological processes, respectively. In plant cells, knowledge of the function of these post-translational modifications under physiological and stress conditions is still very rudimentary. In this addendum, we briefly examine how reactive nitrogen species (RNS) can exert important effects on proteins that could mediate signalling processes in plants.
Introduction
The molecule nitric oxide or nitrogen monoxide (NO) has a long chemical history, but in the last 20 years, it has become the centre of research in many different areas of biology and medicine. In 1772 Joseph Priestley characterized NO as a colourless, no flammable, odourless and toxic gas.Citation1 After the industrial revolution, this compound was only studied as a component of air pollution because NO was involved in the acid rain and in the ozone layer destruction. In 1979, Klepper reported that plants can generate nitric oxide,Citation2 but this publication did not attract the attention of plant researchers until 1987, when nitric oxide was identified as the endothelium-derived relaxing factor (EDRF) by Palmer et al.Citation3 This finding marked the beginning of a new research era in biological systems, as evidenced by the more than 84,000 papers that have been published in the last 20 years concerning nitric oxide biology. In plants, in mid 90s some reports started to remember that plants, as demonstrated by Klepper (1979), could also produce NO, and that this molecule had an ample spectrum of physiological functions as signalling molecule. Although there are still some shadows in the research of NO in plants such as the identification of the enzymatic source/s of NO in physiological and adverse conditions, NO and NO-related molecules are today one of the main research areas in plant physiologyCitation4 because some of the NO effects are due to its direct or indirect interaction with proteins.
Proteins can suffer different post-translational modifications that can affect its activity, conformation, folding, distribution, stability, and therefore its function. These post-translational modifications include formylation, hydroxylation, glycosylation, methylation, oxidation, phosphorylation, ubiquitination, etc. To consider that a certain post-translational modification has physiological significance in cell signalling it must meet several criteria: (i) it must be specific and reversible; (ii) its formation must occur on a physiological time-scale; and (iii) depending on the target, the modification should result in either activation or inhibition.Citation5 Thus, protein phosphorylation is an excellent example of cell signalling.
Nitric oxide (NO) and NO-derived molecules have become one of the most interesting molecules in plant physiology due to the broad spectrum of actions they carry out, including signalling processes. Recently, we reported that salinity induces nitrosative stress in olive plants and we proposed that protein tyrosine nitration could be a good biomarker for nitrosative stress in these plants.Citation6 In this addendum, we describe some additional considerations on the significance of post-translational modifications mediated by reactive nitrogen species (RNS) in the homeostasis of plant cells.
Protein S-nitrosylation
This post-translational modification consists in the binding of a NO group to a cysteine residue of a protein and can alter the function of a broad spectrum of proteins.Citation7 In animal organisms, the S-nitrosylation process is involved in a certain number of patho-physiological situations which can contribute to the generation of nitrosative stress,Citation8 but in plant cells there is much less information on this post-translational modification.Citation9 By proteomics approaches, some putative protein targets for S-nitrosylation have been identified, including cytoskeleton, metabolic, redox-related, stress-related, and signalling/regulating proteins.Citation10 However, only for few proteins there is experimental evidence that they are regulated by this post-translational modification. Thus, in Arabidopsis, nonsymbiotic hemoglobin (AHb1) scavenges NO with production of S-nitrosohemoglobin and reduces NO emission under hypoxic stress.Citation11 Glyceraldehyde-3-phosphate dehydrogenase suffers a reversible inhibition by NO,Citation10 and methionine adenosyltransferase is inhibited by S-nitrosylation.Citation12 The Arabidopsis type-II metacaspase AtMC9 blocks the autoprocessing and activation of AtMC9 zymogen through S-nitrosylation of its catalytic cysteine residue.Citation13 Recently, it has been shown that the S-nitrosylation in the conserved Cys53 of the transcription factor AtMYB2 inhibits its bind to DNA.Citation14 This transcription factor is usually expressed in response to hypoxia, salinity, water stress and ABA. Other potential candidates are the peroxiredoxins which possess one or two active cysteines.
Protein Tyrosine Nitration
This reaction consists in the addition of a nitro group (-NO2) to one of the two equivalent ortho carbons of the aromatic ring of tyrosine residues.Citation15 This post-translational modification seems to be mediated by peroxynitrite (ONOO−),Citation16 a powerful oxidant which is the result of the quick reaction between superoxide radicals (O2• -) and nitric oxide (k = 1.9 × 1010 M −1 s−1).Citation17 In animal cells, protein tyrosine nitration is starting to be used as a biomarker of pathological states and nitrosative stressCitation18–Citation21 because this process can alter the conformation and structure of proteins, the catalytic activity of enzymes, and its susceptibility to proteolysis. Recent experimental data obtained seem to indicate that nitro-tyrosine could also be used as a marker of nitrosative stress in plants.Citation6,Citation23,Citation24 Nevertheless, it must be said that there are also data in animal systems indicating that under physiological conditions some proteins are specifically nitrated.Citation25–Citation27 This suggests that, in some cases, protein tyrosine nitration could be reversible and be involved in signaling.Citation5,Citation18 Thus, it has been described the existence of a nitro-tyrosine denitrase activity which has the capacity to remove specific nitro-tyrosine residues in proteinsCitation22,Citation28 and this process was dependent on NO biosynthesis de novo.Citation5,Citation29,Citation30
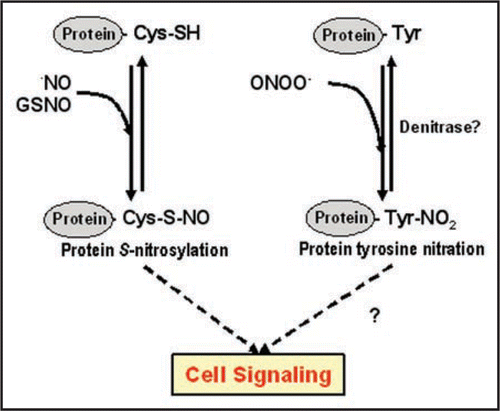
shows a model with the elements described in this addendum considering that the chemical mechanisms are not totally understood. Thus, nitric oxide and GSNO can mediate the S-nitrosylation of proteins, but could S-nitrosoglutathione reductase (GSNOR) catalyze the reverse reaction? Alternatively, protein tyrosine nitration can be induced by peroxinitrite (ONOO−), a process that can be mediated by metalloproteins, peroxidases or transition metals.Citation8 On the other hand, the recently described enzyme nitro-tyrosine denitrase could remove specific nitro-tyrosine residues in proteins and therefore could make this process reversible. These two processes, together or separately, could contribute to cell signalling.
Outlook
The identification of the enzymatic sources of NO in plant cells and its subcellular localization will be essential to identify NO protein targets and their functions. Thus, information on the cell compartmentalization of these post-translational modified proteins could be very important to understand the cross-talk between cell organelles mediated by NO or NO-derived molecules. The identification of a possible nitro-tyrosine denitrase activity in plants could also help evaluate whether this process could contribute to cell signalling like S-nitrosylation.
Figures and Tables
Acknowledgements
This work was supported by grants from the Ministry of Education and Science (BIO2006-14949-C02-01 and BIO2006-14949-C02-02) and Junta de Andalucía (groups CVI 192 and CVI 286).
Addendum to:
References
- Priestley J. Experiments and Observations on Different Kinds of Air 1772; 1 London W. Bowyer and J. Nichols
- Keppler LA. Nitric oxide (NO) and nitrogen dioxide (NO2) emissions from herbicide-treated soybean plants. Atmosperic Environment 1979; 13:537 - 542
- Palmer RM, Ferrige AG, Moncada S. Nitric oxide release accounts for the biological activity of endothelium-derived relaxing factor. Nature 1987; 327:524 - 526
- Neill S, Bright J, Desikan R, Hancock J, Harrison J, Wilson I. Nitric oxide evolution and perception. J Exp Bot 2008; 59:25 - 35
- Koeck T, Stuehr DJ, Aulak KS. Mitochondria and regulated tyrosine nitration. Biochem Soc Trans 2005; 33:1399 - 1403
- Valderrama R, Corpas FJ, Carreras A, Fernández-Ocaña A, Chaki M, Luque F, Gómez-Rodríguez MV, Colmenero-Varea P, del Río LA, Barroso JB. Nitrosative stress in plants. FEBS Letters 2007; 581:453 - 461
- Sun J, Steenbergen C, Murphy E. S-nitrosylation: NO-related redox signaling to protect against oxidative stress. Antioxid Redox Signal 2006; 8:1693 - 1705
- Benhar M, Forrester MT, Stamler JS. Nitrosative stress in the ER: A new role for S-nitrosylation in neurodegenerative diseases. ACS Chemical Biology 2006; 1:355 - 358
- Barroso JB, Corpas FJ, Carreras A, Rodríguez-Serrano M, Esteban FJ, Fernández-Ocaña A, Chaki M, Romero-Puertas MC, Valderrama R, Sandalio LM, del Río LA. Localization of S-nitrosoglutathione and expression of S-nitrosoglutathione reductase in pea plants under cadmium stress. J Exp Bot 2006; 57:1785 - 1793
- Lindermayr C, Saalbach G, Durner J. Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol 2005; 137:921 - 930
- Perazzolli M, Dominici P, Romero-Puertas MC, Zago E, Zeier J, Sonoda M, Lamb C, Delledonne M. Arabidopsis nonsymbiotic hemoglobin AHb1 modulates nitric oxide bioactivity. Plant Cell 2004; 16:2785 - 2794
- Lindermayr C, Saalbach G, Bahnweg G, Durner J. Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. J Biol Chem 2006; 281:4285 - 4291
- Belenghi B, Romero-Puertas MC, Vercammen D, Brackenier A, Inzé D, Delledonne M, Van Breusegem F. Metacaspase activity of Arabidopsis thaliana is regulated by S-nitrosylation of a critical cysteine residue. J Biol Chem 2007; 282:1352 - 1358
- Serpa V, Vernal J, Lamattina L, Grotewold E, Cassia R, Terenzi H. Inhibition of AtMYB2 DNA-binding by nitric oxide involves cysteine S-nitrosylation. Biochem Biophys Res Commun 2007; 361:1048 - 1053
- Gow AJ, Farkouh CR, Munson DA, Posencheg MA, Ischiropoulos H. Biological significance of nitric oxide-mediated protein modifications. Am J Physiol Lung Cell Mol Physiol 2004; 287:L262 - L268
- Radi R. Nitric oxide, oxidants, and protein tyrosine nitration. Proc Natl Acad Sci USA 2004; 101:4003 - 4008
- Kissner R, Nauser T, Bugnon P, Lye PG, Koppenol WH. Formation and properties of peroxynitrite as studied by laser flash photolysis, high-pressure stopped-flow technique, and pulse radiolysis. Chem Res Toxicol 1997; 10:1285 - 1292
- Schopfer FJ, Baker PR, Freeman BA. NO-dependent protein nitration: A cell signaling event or an oxidative inflammatory response?. Trends Biochem Sci 2003; 28:646 - 654
- Ischiropoulos H. Biological selectivity and functional aspects of protein tyrosine nitration. Biochem Biophys Res Commun 2003; 305:776 - 783
- Casoni F, Basso M, Massignan T, Gianazza E, Cheroni C, Salmona M, Bendotti C, Bonetto V. Protein nitration in a mouse model of familial amyotrophic lateral sclerosis: Possible multifunctional role in the pathogenesis. J Biol Chem 2005; 280:16295 - 16304
- Bartesaghi S, Ferrer-Sueta G, Peluffo G, Valez V, Zhang H, Kalyanaraman B, Radi R. Protein tyrosine nitration in hydrophilic and hydrophobic environments. Amino Acids 2007; 32:501 - 515
- Kamisaki Y, Wada K, Bian K, Balabanli B, Davis K, Martin E, Behbod F, Lee YC, Murad F. Protein nitration in a mouse model of familial amyotrophic lateral sclerosis: Possible multifunctional role in the pathogenesis. Proc Natl Acad Sci USA 1998; 95:11584 - 11589
- Corpas FJ, Carreras A, Valderrama R, Chaki M, Palma JM, del Río LA, Barroso JB. Reactive nitrogen species and nitrosative stress in plants. Plant Stress 2007; 1:37 - 41
- Corpas FJ, del Río LA, Barroso JB. Need of biomarkers of nitrosative stress in plants. Trends Plant Sci 2007; 12:436 - 438
- Greenacre SA, Ischiropoulos H. Tyrosine nitration: Localisation, quantification, consequences for protein function and signal transduction. Free Radical Res 2001; 34:541 - 581
- Marcondes S, Turko IV, Murad F. Nitration of succinyl-CoA:3-oxoacid CoA-transferase in rats after endotoxin administration. Proc Natl Acad Sci USA 2001; 98:7146 - 7151
- Tedeschi G, Cappelletti G, Nonnis S, Taverna F, Negri A, Ronchi C, Ronchi S. Tyrosine nitration is a novel post-translational modification occurring on the neural intermediate filament protein peripherin. Neurochem Res 2007; 32:433 - 441
- Irie Y, Saeki M, Kamisaki Y, Martin E, Murad F. Histone H1.2 is a substrate for denitrase, an activity that reduces nitrotyrosine immunoreactivity in proteins. Proc Natl Acad Sci USA 2003; 100:5634 - 5639
- Aulak KS, Koeck T, Crabb JW, Stuehr DJ. Dynamics of protein nitration in cells and mitochondria. Am J Physiol Heart Circ Physiol 2004; 286:H30 - H38
- Heijnen HF, van Donselaar E, Slot JW, Fries DM, Blachard-Fillion B, Hodara R, Lightfoot R, Polydoro M, Spielberg D, Thomson L, Regan EA, Crapo J, Ischiropoulos H. Subcellular localization of tyrosine-nitrated proteins is dictated by reactive oxygen species generating enzymes and by proximity to nitric oxide synthase. Free Radic Biol Med 2006; 40:1903 - 1913