Abstract
The plant hormone cytokinins regulate diverse aspects of plant growth and development. In Arabidopsis, a multi-step TCS system similar to bacterial and yeast TCS is used for cytokinin signaling. In a TCS system, a His sensor kinase perceives the signal by autophosphorylating on a His residue in response to an output signal, and the phosphate group is transferred to a conserved Asp residue in the receiver domain of the response regulator. The response regulator then modulates downstream signaling. Cytokinin multi-step TCS system utilizes an additional component, histidine-containing phosphotransfer domain protein (HPT) to transfer the phosphate group from a sensor kinase to a response regulator in the nucleus. The typical response regulators are classified into either type A or B. The type-B ARRs are transcription activators that act as positive regulators of cytokinin signaling, whereas most of the type-A ARRs are negative regulators of cytokinin signaling. Histidyl-aspartidyl phosphorelays are presumed to be essential for this cytokinin signal transduction in plants. Our studies have shown that ARR7, an A-type response regulator, negatively regulates cytokinin signaling in various aspects by acting as a transcriptional repressor and that the phosphorylation of ARR7 is required for these ARR7-regulated cytokinin-responses. Here I propose potential mechanisms by which the phosphorylation of ARRs is involved in regulating cytokinin-mediated gene expression, mainly based on biochemical and structural studies of bacterial response regulators. Protein-protein interaction and DNA-binding studies using the phosphorylated and the un-phosphorylated forms of the ARR proteins with their structural determination will provide molecular understanding of cytokinin-responsive gene regulation by ARRs.
There are three sensor histidine-kinase cytokinin receptors, CRE1/AHK4/WOL1, AHK2 and AHK3 in Arabidopsis.Citation1 These AHK genes function as positive regulators of cytokinin signaling and possess distinct yet overlapping functions in the regulation of shoot growth, root development, leaf senescence, seed size, germination and cytokinin metabolism.Citation2–Citation4 The AHK2 and AHK3 receptors play prominent roles in the control of leaf development, whereas AHK4 functions in the roots.Citation3 AHK3 is shown to mediate delay in leaf senescence.Citation5 Other His kinases, AHK1, CYTOKININ INDEPENDENT1 (CKI1) and AHK5 (CKI2), function independently of cytokinin action.Citation6–Citation8 AHP1 and AHP2 shuttle between the cytoplasm and the nucleus in a cytokinin-dependent manner.Citation9 Most of AHPs act as redundant, positive regulators of cytokinin signaling except for AHP6.Citation10 AHP6 has a mutation in the conserved histidine residue required to receive the incoming phosphoryl group and thus is pseudo-HPT.Citation11 AHP6 inhibits cytokinin signaling for allowing proxylem formation.Citation12 Twenty-three putative response regulators were identified in Arabidopsis that are classified into type-A or -B.Citation13 The type-B ARRs contain the receiver domain and a large C-terminal region harboring a Myb-like DNA-binding domain and a glutamine-rich region for transcriptional activation and are not inducible by cytokinins.Citation13–Citation15 Cytokinin-inducible type-A ARRs are composed of the receiver domain and a C-terminal extension and act mainly as redundant negative regulators with some functional specificity in cytokinin signaling.Citation16
Presumed autophosphorylation in the AHK receptors in response to cytokinin signal has been demonstrated in CRE1 with in vitro and with a yeast system.Citation17 It was found that CRE1 is not only a kinase that phosphorylates HPTs in the presence of cytokinins but is also a phosphatase that dephosphorylates HPTs in the absence of cytokinins. This bidirectional phosphorelay network ensures that in the absence of cytokinins, the signaling pathway is quickly inactivated. AHP1, AHP2 and AHP3 are shown phospho-transmitting proteins.Citation18 Various studies have shown that both B-type ARR and A-type ARR can acquire phosphoryl group from the AHP proteins in vitro.Citation19,Citation20 We have shown that the Asp-85 mutation of ARR7 into the Asp residue blocked the phosphorylation of ARR7 by the Arabidopsis protein extracts in vitro, and abolished the ability of ARR7 to suppress root-elongation inhibition, callus formation and cytokinin-inducible gene expression in transgenic Arabidopsis.Citation21 Conversely, overexpression of ARR7D85E harboring the active phosphorylated state in transgenic Arabidopsis caused severe effects on the function of the shoot apical meristem, including meristems arrested for several days after expansion of the cotyledons that resulted in an almost complete block of organ formation.Citation22 GeneChip analysis has shown that ARR7 mainly acts as a transcriptional repressor for a variety of early cytokinin-regulated genes encoding transcription factors, signal transmitters, plant development and cellular metabolism.Citation23 Thus the phosphorylation of ARR7 is critical for its function as a negative regulator of various cytokinin-signaling pathways in Arabidopsis plants. Overexpression of ARR2D80E in transgenic Arabidopsis caused severe developmental defects, but the transgenic Arabidopsis plants overexpressing ARR2D80N did not exhibit ARR2-mediated delay in senescence,Citation5 suggesting that the phosphorylation of B-type ARR also plays a critical role in cytokinin signal transduction.
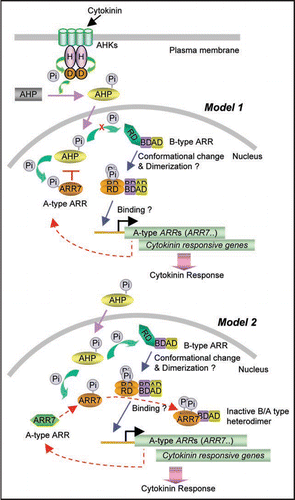
The A-type ARR proteins including ARR7 were shown to interact with various AHP proteins in the yeast two-hybrid assays and in vitro binding assays.Citation24 The same study showed that A-type ARRs do not interact with B-type ARRs. Based on these results, the authors proposed that squelching of AHP proteins by A-type ARRs through protein-protein interaction interferes AHP-mediated signaling, resulting in negative regulation of cytokinin signaling as depicted in model 1 of . Alternative possibility is that only the phosphorylated form of the A-type ARR proteins might bind the B-type ARR proteins through protein-protein interaction mediated by their N-terminal receiver domains phosphorylated, resulting in inhibition of B-type ARR function (model 2 of ). This model is principally based on biochemical and structural studies of the bacterial response regulators.Citation25–Citation28 Bacterial response regulators undergo large conformational changes upon phosphorylation on a conserved Asp residue in the receiver domain, forming dimers that allow DNA target site recognition and transcriptional activation. Likewise, the B-type ARR proteins might undergo conformational changes upon phosphorylation of the receiver domain to form functional dimers that bind to the promoter regions of the cytokinin-inducible genes including A-type ARR genes. This hypothesis is backed up by the observation that the transactivating function of ARR1 was masked by its receiver domain in transient expression experimentsCitation14 and transgenic Arabidopsis overexpressing truncated ARR1 missing the receiver domain exhibited a phenotypic change whereas full-length ARR1 did not display phenotypic change.Citation29 The A-type ARR proteins will be accumulated and the AHP proteins will phosphorylate them. Because both B-type ARRs and A-type ARRs contain the highly conserved receiver domains, the phosphorylated A-type ARR proteins might interact with the phosphorylated B-type ARR proteins through their receiver domains, forming inactive forms of the B/A-type ARR heterodimers. Accumulation of the inactive B/A-type ARR heterodimers will eventually result in inhibition of cytokinin-mediated gene expression. Our study with cycloheximide-chase assays in Arabidopsis mesophyll protoplasts showed that the half-life of LUC:ARR7 is 60 min, while the LUC:NLS protein remains stable.Citation21 MG132 inhibited the degradation of ARR7:LUC to some degree in the presence of cycloheximide. These results suggest that ARR7 is labile and subject to proteasome-mediated degradation, in part. The same assays showed that LUC:ARR7D85N has 77 min of half-life, suggesting that phosphorylation itself may not have a role in modulating ARR7 protein stability. When cytokinin levels decline, CRE1 will dephosphorylate AHPs by its intrinsic phosphatase activity,Citation17 shutting off cytokinin signal transduction. The A-type ARR proteins will be removed because of their relatively fast degradation kinetics. The repression of the B-type ARR proteins by the A-type ARR proteins will then be released. Moreover, the phosphate groups on the B-type ARR proteins have very short half-life.Citation19 Therefore, the phosphorylated B-type ARR proteins will lose their phosphate groups and be ready for the next round of cytokinin action. I believe that his model is testable as being proven for bacterial response regulators by using biochemical approaches including protein-protein interaction and DNA-binding studies along with their structural determination.
Abbreviations
| AHK | = | arabidopsis histidine kinase |
| AHP | = | arabidopsis histidine-containing phosphotransfer domain protein |
| ARR | = | arabidopsis response regulator |
| TCS | = | two-component signaling |
Figures and Tables
Figure 1 Working models on the roles of the phosphorylation of the ARR proteins in cytokinin-responsive gene regulation in Arabidopsis. In cytokinin signaling, the receiver domain of B-type ARRs is phosphorylated by AHPs that acquire the phosphoryl groups from the AHK sensor kinases. It is hypothesized that B-type ARRs undergo conformational changes upon phosphorylation on the receiver domain, allowing formation of a functional dimer of B-type ARR that binds to the promoter regions of the cytokinin-inducible genes including A-type ARR genes. Model 1 proposes squelching mechanism of AHPs by phosphorylated ARRs, preventing interaction between AHPs and B-type ARRs. Model 2 proposes formation of the inactive heterodimer of B-type ARR and A-type ARR by protein-protein interaction mediated by their highly conserved receiver domains that are phosphorylated. Refer to text for detailed descriptions. Red dotted lines indicate negative feedback inhibitory pathway. RD, receiver domain; BD, DNA binding domain; AD, transcriptional activation domain; H, histidine residue for accepting phosphoryl group; D, aspartic acid residue for accepting phosphoryl group; Pi, phosphoryl goup.
Acknowledgements
This work was supported by grants from the Agricultural Plant Stress Research Center (R11-2001-092-04001-0) funded by the Korea Science and Engineering Foundation and the Plant Diversity Research Center of 21st Century Frontier Research Program (PF06302-01) funded by the Ministry of Science and Technology of the Korean Government to J. Kim.
Addendum to:
References
- Heyl A, Schmülling T. Cytokinin signal perception and transduction. Curr Opin Plant Biol 2003; 6:480 - 488
- Higuchi M, Pischke MS, Mähönen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S. In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 2004; 8:8821 - 8826
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 2004; 16:1365 - 1377
- Riefler M, Novak O, Strnad M, Schmülling T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth leaf senescence seed size germination root development and cytokinin metabolism. Plant Cell 2006; 18:40 - 54
- Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I. Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc Natl Acad Sci USA 2006; 103:814 - 819
- Urao T, Yakubov B, Satoh R, Yamaguchi Shinozaki K, Seki M, Hirayama T, Shinozaki K. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 1999; 11:1743 - 1754
- Pischke MS, Jones LG, Otsuga D, Fernandez DE, Drews GN, Sussman MR. An Arabidopsis histidine kinase is essential for megagametogenesis. Proc Natl Acad Sci USA 2002; 99:15800 - 15805
- Iwama A, Yamashino T, Tanaka Y, Sakakibara H, Kakimoto T, Sato S, Kato T, Tabata S, Nagatani A, Mizuno T. AHK5 histidine kinase regulates root elongation through an ETR1-dependent abscisic acid and ethylene signaling pathway in Arabidopsis thaliana. Plant Cell Physiol 2007; 48:375 - 380
- Hwang I, Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 2001; 413:383 - 389
- Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, Maxwell BB, Perdue TD, Schaller GE, Alonso JM, Ecker JR, Kieber JJ. The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 2006; 18:3073 - 3087
- Müller B, Sheen J. Advances in cytokinin signaling. Science 2007; 318:68 - 69
- Mähönen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, Tormakangas K, Ikeda Y, Oka A, Kakimoto T, Helariutta Y. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 2006; 311:94 - 98
- Hwang I, Chen HC, Sheen J. Two-component signal transduction pathways in Arabidopsis. Plant Physiol 2002; 129:500 - 515
- Sakai H, Aoyama T, Oka A. Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J 2000; 24:703 - 711
- Hosoda K, Imamura A, Katoh E, Hatta T, Tachiki M, Yamada H, Mizuno T, Yamazaki T. Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 2002; 14:2015 - 2029
- To JPC, Haberer G, Ferreira FJ, Deruère J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 2004; 16:658 - 671
- Mähönen AP, Higuchi M, Törmäkangas K, Miyawaki K, Pischke MS, Sussman MR, Helariutta Y, Kakimoto T. Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr Biol 2006; 16:1116 - 1122
- Suzuki T, Imamura A, Ueguchi C, Mizuno T. Histidine-containing phosphotransfer (HPt) signal transducers implicated in His-to-Asp phosphorelay in Arabidopsis. Plant Cell Physiol 1998; 39:1258 - 1268
- Imamura A, Kiba T, Tajima Y, Yamashino T, Mizuno T. In vivo and in vitro characterization of the ARR11 response regulator implicated in the His-to-Asp phosphorelay signal transduction in Arabidopsis thaliana. Plant Cell Physiol 2003; 44:122 - 131
- Kiba T, Aoki K, Sakakibara H, Mizuno T. Arabidopsis response regulator ARR22 ectopic expression of which results in phenotypes similar to the wol cytokinin-receptor mutant. Plant Cell Physiol 2004; 45:1063 - 1077
- Lee DJ, Kim S, Ha YM, Kim J. Phosphorylation of Arabidopsis response regulator 7 (ARR7) at the putative phospho-accepting site is required for ARR7 to act as a negative regulator of cytokinin signaling. Planta 2008; 227:577 - 587
- Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 2005; 438:1172 - 1175
- Lee DJ, Park JY, Ku SJ, Ha YM, Kim S, Kim MD, Oh MH, Kim J. Genome-wide expression profiling of ARABIDOPSIS RESPONSE REGULATOR 7(ARR7) overexpression in cytokinin response. Mol Genet Genomics 2007; 277:115 - 137
- Dortay H, Mehnert N, Burkle L, Schmülling T, Heyl A. Analysis of protein interactions within the cytokinin-signaling pathway of Arabidopsis thaliana. FEBS J 2006; 273:4631 - 4644
- Birck C, Mourey L, Gouet P, Fabry B, Schumacher J, Rousseau P, Kahn D, Samama JP. Conformational changes induced by phosphorylation of the FixJ receiver domain. Structure 1999; 7:1505 - 1515
- Birck C, Malfois M, Svergun D, Samama JP. Insights into signal transduction revealed by the low resolution structure of the FixJ response regulator. J Mol Biol 2002; 321:447 - 457
- Lewis RJ, Scott DJ, Brannigan JA, Ladds JC, Cervin MA, Spiegelman GB, Hoggett JG, Barak I, Wilkinson AJ. Dimer formation and transcription activation in the sporulation response regulator Spo0A. J Mol Biol 2002; 316:235 - 245
- Maris AE, Sawaya MR, Kaczor Grzeskowiak M, Jarvis MR, Bearson SM, Kopka ML, Schroder I, Gunsalus RP, Dickerson RE. Dimerization allows DNA target site recognition by the NarL response regulator. Nat Struct Biol 2002; 9:771 - 778
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A. ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 2001; 294:1519 - 1521 (2001)