Abstract
Allelopathic interaction between plants is thought to involve the release of phytotoxic allelochemicals by one species, thus inhibiting the growth of neighboring species in competition for limited resources. Sorgoleone represents one of the more potent allelochemicals characterized to date, and its prolific production in root hair cells of Sorghum spp. has made the investigation of its biosynthetic pathway ideally-suited for functional genomics investigations. Through the use of a recently-released EST data set generated from isolated Sorghum bicolor root hair cells, significant inroads have been made toward the identification of genes and the corresponding enzymes involved in the biosynthesis of this compound in root hairs. Here we provide additional information concerning our recent report on the identification of a 5-n-alk(en)ylresorcinol utilizing O-methyltransferase, as well as other key enzymes likely to participate in the biosynthesis of this important allelochemical.
Introduction
Allelopathic interactions have been proposed to have profound effects on the evolution of natural ecosystems by conferring a competitive advantage to allelochemical-producing species, or species resistant to interference from a given allelochemical.Citation1,Citation2 In addition, allelochemical release by certain crop species such as barley, rye and sorghum is thought to play a significant role in their utility as weed suppressants when used as cover crops.Citation3,Citation4 Certain sorghum species such as Sudangrass (Sorghum sudanense) can reportedly produce largely weed-free monocultures without the use of synthetic herbicides.Citation5
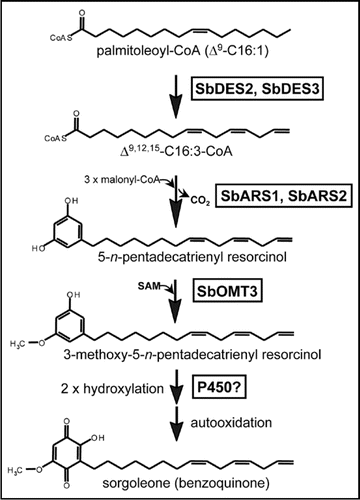
Current evidence suggests that a highly bioactive benzoquinone synthesized in root hair cells of Sorghum spp. termed sorgoleone (2-hydroxy-5-methoxy-3-[(Z,Z)-8′,11′,14′-pentadecatriene]-p-benzoquinone—Fig. 1) may represent the principal chemical constituent responsible for conferring allelopathic properties.Citation6–Citation8 Studies performed in vitro with purified sorgoleone have demonstrated its effectiveness as a broad-spectrum plant growth inhibitor active against many agronomically important monocot and dicot weed species at micromolar concentrations. In fact, this property, coupled with its prolonged half-life in soil and potentially complex mode of action, has led to the suggestion that sorgoleone could be developed as a useful natural product alternative to synthetic herbicides.Citation6–Citation12 Interestingly, within the Poaceae (grass) family, sorgoleone biosynthesis may be restricted to members of the genus Sorghum, as a limited survey involving root samples collected from 17 different Poaceae accessions failed to detect sorgoleone production even in the most closely-related Panicoideae sub-family members ().
Use of Functional Genomics to Explore Sorgoleone Biosynthesis
As mentioned, sorgoleone biosynthesis appears to occur exclusively in root hair cells, which in sorghum appear as cytoplasmically dense cells filled with numerous osmiophilic deposits presumably associated with the rhizosecretion of sorgoleone, which can constitute as much as 85% of the exudate dry weight in some cultivars.Citation8,Citation13 The cell-specific localization and prolific output of the sorgoleone biosynthetic pathway rendered the use of expressed sequence tag (EST) analysis the obvious method of choice in our efforts to isolate genes encoding sorgoleone biosynthetic enzymes.Citation14 Labeling studies performed by Fate & Lynn,Citation15 first demonstrated that biosynthesis proceeds through the action of an alkylresorcinol synthase (ARS), a novel type III polyketide synthase activity utilizing fatty acyl-CoA starter units (). Subsequently, both the predicted 5-n-pentadecatrienyl resorcinol as well as a 3-methyl ether derivative of this compound were identified in sorghum root extracts, indicating that dihydroxylation of the resorcinol ring is preceded by O-methylation at the 3-hydroxyl position.Citation14,Citation16 The resulting chemically unstable hydroquinone rapidly oxidizes to the bioactive sorgoleone benzoquinone once released into the rhizosphere, where it may persist in soil for extended periods.Citation7,Citation8,Citation17 De novo synthesis of sorgoleone from available palmitoleoyl-CoA, would therefore likely require, at a minimum, the participation of a ΔCitation12 and ΔCitation15 fatty acid desaturase (DES), ARS, a 3-O-methyltransferase (OMT), and a cytochrome P450 ().
To obtain candidate sequences for each of these proposed enzyme classes, 5,468 quality 5′-sequenced ESTs were generated from a primary phagemid cDNA library prepared from isolated root hair cells, and all sequence data has been recently deposited in GenBank. Importantly, the S. bicolor genotype BTx623 was used as the source of the root hair tissues, therefore the ESTs can be directly compared to the emergent S. bicolor whole-genome sequence which will also be derived from BTx623.Citation18 Clustering and annotation of the root hair data set revealed 7 unique DES-like sequences, 5 polyketide synthase (PKS)-like sequences, 12 OMT-like sequences, and 11 P450-like sequences, thus all of the required enzyme classes proposed for sorgoleone biosynthesis were represented. Using Saccharomyces cerevisiae as a heterologous host, we have recently demonstrated the ability of two fatty acid desaturases (SbDES2, SbDES3) identified from the root hair EST data set to catalyze the formation of the unusual 16:3Δ9,12,15 fatty acyl-CoA biosynthetic precursor () in vivo, strongly suggesting they perform this same function in planta.Citation19 The functional characterization of an OMT proposed to perform the 3-O-methylation of the 5-n-pentadecatrienyl resorcinol intermediate, designated SbOMT3, is described in our recent report which also provides secondary analyses of the root hair EST data.Citation14 Two PKS-like sequences identified from the root hair ESTs, tentatively designated SbARS1 and SbARS2 (), have also recently been shown to catalyze the formation of 5-n-alk(en)ylresorcinols utilizing an array of saturated and unsaturated fatty acyl-CoA starter units, thus representing the first ARS-type enzymes cloned from higher plants (manuscript in preparation).
Potential Genetic Redundancy of the 3-O-Methylation Function
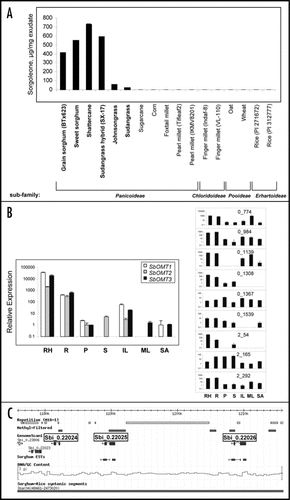
Recombinant enzyme studies involving three of the OMT-like enzymes (SbOMT1, SbOMT2, SbOMT3—) identified among the root hair ESTs were described in our recent report,Citation14 and SbOMT3 demonstrated a marked preference for alkylresorcinolic substrates among a panel of benzene derivatives tested in that study. As mentioned, in total 12 unique sequences resembling OMTs were identified in root hairs, and expression patterns in different sorghum tissues were determined for all of these (). In addition to SbOMT1, SbOMT2, and SbOMT3, two of the remaining 9 OMT-like contig sequences (2_54 and 0_1308; ) were found to be expressed predominantly in root hairs, and were also among the most highly expressed sequences in this cell type.Citation14 Using the partial sequence data available from contig 2_54, a full length open reading encoding an approximately 40.7 kDa OMT-like protein could be deduced when aligned with the available sorghum genome draft sequence. Interestingly 2_54 shares 93% amino acid identity with SbOMT3, and all of critical active site residues proposed for SbOMT3 (based on molecular modeling studies)Citation14 appear to be conserved, suggesting the possibility that this putative OMT could have similar catalytic properties. While the current sorghum genome assembly (Sorbi0; http://www.phytozome.net/sorghum) is a preliminary draft based on a partial data set, available evidence indicates that SbOMT3 is actually represented as three closely-linked copies, one of which is an apparent 5′-truncated pseudogene (). Of the three predicted OMT-like genes residing within an approximately 30 kb chromosomal region, Sbi_0.22025 and Sbi_0.22026 share 100% and 99% nucleotide identity with the SbOMT3 open reading frame predicted from the root hair ESTs, respectively. The truncated copy corresponds to Sbi_0.22025 (), and lacks the first approximately 500 bp of the OMT open reading frame, with the remainder sharing 98% identity with SbOMT3. Extensive sequence identity also exists within the regions flanking the reading frames of Sbi_0.22024, Sbi_0.22025 and Sbi_0.22026, suggesting the occurrence of a recent gene duplication event. The predicted gene corresponding to root hair contig 2_54 resides on an entirely different Sorbi0 super-cluster and may not be closely-linked to SbOMT3 (Sbi_0.22025). While the high degree of sequence identity shared between the 5′ and 3′ flanking regions of SbOMT3 and Sbi_0.22026 would suggest identical expression patterns for the two genes, even minor changes within critical motifs could lead to significant alterations in promoter activity, potentially providing the basis for functional divergence. Examining the substrate specificity of putative OMT 2_54, and the potential role played by Sbi_0.22026 in sorgoleone biosynthesis represent just two out of many intriguing questions awaiting further exploration.
Figures and Tables
Figure 2 Survey of sorgoleone production in grasses, expression patterns of O-methyltransferase-like sequences identified in root hairs, and genomic organization of SbOMT3-like copies are shown. (A) Sorgoleone contents were determined in root extracts prepared from representative Poaceae accessions by GC-MS analysis;Citation14,Citation20 including grain sorghum (S. bicolor ‘BTx623’), sweet sorghum (S. bicolor ‘Della’), shattercane (S. bicolor ssp. drummondii), sorghum-sudangrass hybrid (S. bicolor × sudanense ‘SX-17’), johnsongrass (S. halapense L. Pres.), sudangrass (S. sudanense ‘Excel’), sugarcane (Saccharum spp. hybrid ‘HoCP 91-555’), corn (Zea mays ‘Merit’), foxtail millet (Setaria italica ‘Golden German’), pearl millet (Pennisetum glaucum ‘Tifleaf2’ and ‘IKMV8201’), finger millet (Eleusine coracana ‘Indaf-8’ and ‘VL-110’), Oat (Avena sativa ‘Bob’), wheat (Triticum aestivum ‘Coker 9553’) and two allelopathic rice varieties (Oryza sativa ‘PI271672’ and ‘PI312777’)Citation21. Averages obtained from replicate samples are shown, and associated Poaceae subfamilies are indicated by brackets as assigned by Kellogg.Citation22 (B) Relative expression patterns in different sorghum tissues for OMT-like sequences identified in root hairs were determined by quantitative real-time RT-PCR. SbOMT1-3 graph (shown at left) was adapted from Baerson et al.;Citation14 contig I.D. numbers for the remaining 9 OMT-like sequences identified (shown at right) are provided within each graph. RH, root hair; R, total root; P, panicle; S, stem; IL, immature leaf; ML, mature leaf; SA, shoot apex. (C) Sorghum Genome Browser return (http://www.phytozome.net) is shown indicating three closely-linked SbOMT3-like copies within the S. bicolor (‘BTx623’) genome (corresponding to GenomeScan1 predicted ORFs Sbi_0.22024, Sbi_0.22025 and Sbi_0.22026 shown in figure).
Addendum to:
References
- Schulz M, Wieland I. Variation in metabolism of BOA among species in various field communities—biochemical evidence for co-evolutionary processes in plant communities?. Chemoecology 1999; 9:133 - 141
- Bais HP, Park SW, Weir TL, Callaway RM, Vivanco JM. How plants communicate using the underground information superhighway. Trends Plant Sci 2004; 9:26 - 32
- Duke SO, Rimando AM, Baerson SR, Scheffler BE, Ota E, Belz RG. Strategies for the use of natural products for weed management. J Pestic Sci 2002; 27:298 - 306
- Weston LA, Duke SO. Weed and Crop Allelopathy. Crit Rev Plant Sci 2003; 22:367 - 389
- Duke SO, Belz RG, Baerson SR, Pan Z, Cook DD, Dayan FE. The potential for advances in crop allelopathy. Outlooks Pest Management 2005; 16:64 - 68
- Netzly DM, Butler LG. Roots of Sorghum exude hydrophobic droplets containing biologically active components. Crop Sci 1986; 26:775 - 778
- Einhellig FA, Souza IF. Phytotoxicity of sorgoleone found in grain sorghum root exudates. J Chem Ecol 1992; 18:1 - 11
- Czarnota MA, Paul RN, Dayan FE, Nimbal CI, Weston LA. Mode of action, localization of production, chemical nature, and activity of sorgoleone: a potent PSII inhibitor in Sorghum spp. root exudates. Weed Technol 2001; 15:813 - 825
- Nimbal CI, Pedersen JF, Yerkes CN, Weston LA, Weller SC. Phytotoxicity and distribution of sorgoleone in grain sorghum germplasm. J Agric Food Chem 1996; 44:1343 - 1347
- Rimando AM, Dayan FE, Czarnota MA, Weston LA, Duke SO. A new photosystem II electron transfer inhibitor from Sorghum bicolor. J Nat Prod 1998; 61:927 - 930
- Bertin C, Yang X, Weston LA. The role of root exudates and allelochemicals in the rhizosphere. Plant Soil 2003; 256:67 - 83
- Duke SO. Weeding with transgenes. Trends Biotechnol 2003; 21:192 - 195
- Czarnota MA, Paul RN, Weston LA, Duke SO. Anatomy of sorgoleone-secreting root hairs of Sorghum species. Int J Plant Sci 2003; 164:861 - 866
- Baerson SR, Dayan FE, Rimando AM, Nanayakkara NP, Liu CJ, Schröder J, Fishbein M, Pan Z, Kagan IA, Pratt LH, Cordonnier-Pratt MM, Duke SO. A functional genomics investigation of allelochemical biosynthesis in Sorghum bicolor root hairs. J Biol Chem 2008; 283:3231 - 3247
- Fate GD, Lynn DG. Xenognosin methylation is critical in defining the chemical potential gradient that regulates the spatial distribution in striga pathogenesis. J Amer Chem Soc 1996; 118:11369 - 11376
- Dayan FE, Kagan IA, Rimando AM. Elucidation of the biosynthetic pathway of the allelochemical sorgoleone using retrobiosynthetic NMR analysis. J Biol Chem 2003; 278:28607 - 28611
- Netzly DM, Riopel JL, Ejeta G, Butler LG. Germination stimulants of witchweed (Striga asiatica) from hydrophobic root exudate of sorghum (Sorghum bicolor). Weed Sci 1988; 36:441 - 446
- Paterson AH. A cornucopia of genomes. Genome Biol 2006; 7:311
- Pan Z, Rimando AM, Baerson SR, Fishbein M, Duke SO. Functional characterization of desaturases involved in the formation of the terminal double bond of an unusual 16:3Δ9,12,15 fatty acid isolated from Sorghum bicolor root hairs. J Biol Chem 2007; 282:4326 - 4335
- Rimando AM, Kagan IA, Dayan FE, Czarnota MA, Weston LA. Petroski RJ, Tellez MR, Behle RW. Chemical basis for weed suppressive activity of sorghum. Semiochemicals in Pest and Weed Control 2005; Washington DC American Chemical Society 59 - 70
- Dilday RH, Yan WG, Moldenhauer KAK, Gravois KA. Olofsdotter M. Allelopathic activity in rice for controlling major aquatic weeds. Allelopathy in Rice 1998; Philippines International Rice Research Institute 7 - 26
- Kellogg EA. Relationships of cereal crops and other grasses. Proc Natl Acad Sci USA 1998; 101:2040 - 2045