Abstract
Peroxisomes are subcellular organelles with multiple functions mediated by their plasticity and dynamic behavior in plants. Changes in their shape, size, number and enzyme content occur in response to developmental and metabolic cues as well as environmental conditions. The number of peroxisomes per cell is thus mainly determined by its capacity to proliferate. In mammals, peroxisome proliferators such as the hypolipidemic drug clofibrate are perceived by the Peroxisome Proliferator-Activated Receptors (PPARs) nuclear receptors. Therein, activated transcription of the peroxisome biogenesis PEX11 genes and the recruitment of dynamin-related proteins lead to peroxisome proliferation. We recently reported that Arabidopsis thaliana, despite of lacking a PPAR homolog protein, activated the proliferation of peroxisomes in response to clofibrate. Concomitantly, clofibrate activated the expression of wound-responsive genes through the jasmonic acid signaling master regulator COI1 F-box protein. Besides, wounding activated the expression of the peroxisome biogenesis-related PEX1 and PEX14 genes, but not of PEX11 or DRP3A, which analogously to mammals, code for the main regulators of peroxisome proliferation in Arabidopsis. Thus, wounding did not activate peroxisome proliferation. Noteworthy, jasmonic acid-treated plants contained fewer but larger peroxisomes. Despite of the cross-talk between clofibrate- and wound-induced signaling, the proliferation of peroxisomes and the wound-activated defense remained uncoupled.
Increasing evidence that peroxisomes participate in developmental as well as defense-related processes in plants has placed this organelle as a main focus of many plant biology researchers. Although the function of peroxisomes was linked since their discovery to the β-oxidation-mediated degradation of fatty acids,Citation1 we now know that peroxisomes also participate in other basic processes of plants, such as the photorespiratory glycolate pathwayCitation2 and the glyoxylate cycle,Citation3 as well as in the generation of molecules with potential signal activity.Citation4 Among these plant signaling molecules, jasmonatesCitation5, salicylatesCitation6 and the auxin indole3-acetic acid (IAA)Citation7 have all in common their peroxisomal β-oxidation requirement. Moreover, peroxisomes have an essential role in the production and degradation of oxygen and nitrogen reactive species, whose signaling potential is extensively supported in different living organisms.Citation8,Citation9 As a result of the signal-generating function of peroxisomes, these organelles participate in the regulation of many developmental processes of plants including photomorphogenesis,Citation10 embryogenesis,Citation11 seed production,Citation12,Citation13 germination and early postgerminative growth,Citation14,Citation15 floweringCitation16 and leaf senescence.Citation17 Moreover, substantial evidence for a role of leaf peroxisomes in defense against pathogens and herbivores has been recently supported by proteomic analysis.Citation18
However, despite the increasing evidences of different functions for peroxisomes, very little is known about the signaling pathways involved as well as the requirement of dynamic changes in morphology and number of peroxisomes. In yeast and mammals, many of the peroxisome functions are tightly linked to changes in either endogenous metabolic conditions or external aggressions that trigger the proliferation of the organelle. This process, which is naturally triggered by endogenous lipidic signals named as peroxisome proliferators can be also activated by molecules that mimic the endogenous signals. Among the synthetic ligands, the hipolipidemic drugs fibrates and the insulin sensitizers thiazolidinediones are PPARα and PPARγ agonists, respectively, which have been extensively used as therapeutic agents in human pathologies. Endogenous or synthetic peroxisome proliferators act as ligands of nuclear receptors of the Peroxisome Proliferator-Activated Receptors (PPARs), which function indeed as endogenous metabolic sensors.Citation19 The three classes of PPARs, PPARα, PPARβ/δ and PPARγ, are ligand-activated transcription factors of the nuclear hormone receptor superfamily.Citation20 Nevertheless, the functions of PPARs in mammals go far beyond their involvement as regulators of peroxisome proliferation and regulate many other processes in association to other nuclear receptors and coregulators.
For their functions as regulators of peroxisome proliferation, PPARs activate the transcription of genes coding for the peroxisome biogenesis PEX11 protein family and promote the participation of dynamin-related proteins during peroxisome growth, division and proliferation.Citation21
Plant cells have also the capacity to change their peroxisome population either in response to stress factorsCitation22,Citation23 or treatment with the synthetic peroxisome proliferator clofibrate.Citation24 However, the comparison of plants to mammals indicates that although PEX11 and the dynamin-related DRP3A are conserved components of the pathway required to trigger peroxisome proliferation,Citation25–Citation27 the known genome of plants do not include any gene coding for a PPAR homolog protein (www.arabidopsis.org; http://www.tigr.org/tdb/e2k1/osa1/). Nevertheless, plant cells seem to contain molecules with the capacity to act as ligands of PPARs as well as the signaling components necessary to sustain the subsequent peroxisome of proliferation, as demonstrated by expression of a PPARα gene from Xenopus in tobacco plants.Citation28 We have recently reported that clofibrate-induced peroxisome proliferation in Arabidopsis is accompanied by the COI1-mdiated activation of wound- and jasmonic acid (JA)-activated signaling, and also that wounding activated the expression of peroxisome biogenesis genes such as PEX1 and PEX14 but not PEX11 gene family or DRP3A gene.Citation24 These data suggest that there is a significant cross-regulation of clofibrate- and wound-activated signaling. However, these regulatory interactions do not support a functional connection between wound-related defense and peroxisome proliferation as both processes remained uncoupled in Arabidopsis.
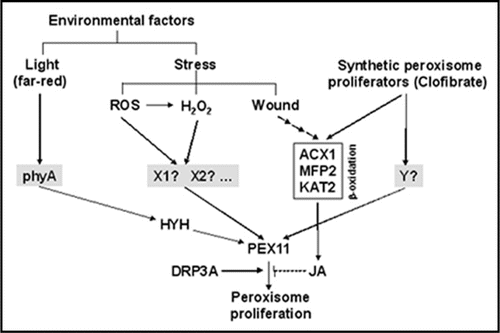
It is noteworthy to mention that JA, a central positive regulator of wound-activated defense, behaved as a negative regulator of peroxisome proliferation as demonstrated by the reduced peroxisome content per cell in leaves from JA treated plants.Citation24 JA negative effect may represent indeed a regulatory loop to control peroxisome proliferation in response to clofibrate (). Although the information available on the components involved in the perception and further signaling of peroxisome proliferators in plants is still very limited, recent data have shown that light activates the proliferation of peroxisomes during seedling photomorphogenesis.Citation29 It required the perception by phytochrome A and the upregulation of PEX11b gene mediated by the bZIP transcription factor HY5 Homolog HYH.Citation29
Altogether, recent findings suggest that the proliferation of peroxisomes in plants, although conserved in the components functioning in the late steps of the pathway, uses multiple perception systems depending on the triggering factor. The identification of the different plant perception systems working as the corresponding PPARs in mammals will be essential in the near future to understand the function of peroxisome proliferation in plant growth, development and defense.
Figures and Tables
Figure 1 Proliferation of peroxisomes triggered by environmental and synthetic proliferators in Arabidopsis. ROS, reactive oxygen species; ACX1; acyl-CoA oxidase 1; MFP2, multifunctional protein 2; KAT2, 3-ketoacyl-CoA thiolase 2; HYH, HY5 Homolog; DRP3A, dynamin-related protein 3A; JA, jasmonic acid; phyA, phytochrome A. The components highlighted in grey represent the mostly unknown potential specific receptors of different peroxisome proliferators. The solid lines indicate positive effect whereas dashed lines represent negative modulation. Recent findings by Castillo et al., (2008)Citation24 and Desai and Hu (2008)Citation29 are integrated in this scheme.
Addendum to:
References
- Goepfert S, Poirier Y. Beta-oxidation in fatty acid degradation and beyond. Curr Opin Plant Biol 2007; 10:245 - 251
- Reumann S, Weber AP. Plant peroxisomes respire in the light: some gaps of the photo-respiratory C2 cycle have become filled—others remain. Biochim Biophys Acta 2006; 1763:1496 - 1510
- Kunze M, Pracharoenwattana I, Smith SM, Hartig A. A central role for the peroxisomal membrane in glyoxylate cycle function. Biochim Biophys Acta 2006; 1763:1441 - 1452
- Nyathi Y, Baker A. Plant peroxisomes as a source of signalling molecules. Biochim Biophys Acta 2006; 1763:1478 - 1495
- Castillo MC, Martínez C, Buchala A, Métraux JP, León J. Gene-specific involvement of beta-oxidation in wound-activated responses in Arabidopsis. Plant Physiol 2004; 135:85 - 94
- Jarvis AP, Schaaf O, Oldham NJ. 3-hydroxy-3-phenylpropanoic acid is an intermediate in the biosynthesis of benzoic acid and salicylic acid but benzaldehyde is not. Planta 2000; 212:119 - 126
- Zolman BK, Yoder A, Bartel B. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 2000; 156:1323 - 1337
- Schrader M, Fahimi HD. Peroxisomes and oxidative stress. Biochim Biophys Acta 2006; 1763:1755 - 1766
- Corpas FJ, Barroso JB, del Río LA. Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant Sci 2001; 6:145 - 150
- Hu J, Aguirre M, Peto C, Alonso J, Ecker J, Chory J. A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science 2002; 297:405 - 409
- Sparkes IA, Brandizzi F, Slocombe SP, El-Shami M, Hawes C, Baker A. An Arabidopsis pex10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol 2003; 133:1809 - 1819
- Fan J, Quan S, Orth T, Awai C, Chory J, Hu J. The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol 2005; 139:231 - 239
- Footitt S, Dietrich D, Fait A, Fernie AR, Holdsworth MJ, Baker A, Theodoulou FL. The COMATOSE ATP-binding cassette transporter is required for full fertility in Arabidopsis. Plant Physiol 2007; 144:1467 - 1480
- Footitt S, Marquez J, Schmuths H, Baker A, Theodoulou FL, Holdsworth M. Analysis of the role of COMATOSE and peroxisomal beta-oxidation in the determination of germination potential in Arabidopsis. J Exp Bot 2006; 57:2805 - 2814
- Eastmond PJ, Germain V, Lange PR, Bryce JH, Smith SM, Graham IA. Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc Natl Acad Sci USA 2000; 97:5669 - 5674
- Richmond TA, Bleecker AB. A defect in beta-oxidation causes abnormal inflorescence development in Arabidopsis. Plant Cell 1999; 11:1911 - 1924
- Hayashi M, Toriyama K, Kondo M, Kato A, Mano S, De Bellis L, Hayashi-Ishimaru Y, Yamaguchi K, Hayashi H, Nishimura M. Functional transformation of plant peroxisomes. Cell Biochem Biophys 2000; 32:295 - 304
- Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, Antonicelli GE, Rasche N, Lüder F, Weckwerth W, Jahn O. Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 2007; 19:3170 - 3193
- Grimaldi PA. Peroxisome proliferator-activated receptors as sensors of fatty acids and derivatives. Cell Mol Life Sci 2007; 64:2459 - 2464
- Michalik L, Auwerx J, Berger JP, Chatterjee VK, Glass CK, Gonzalez FJ, Grimaldi PA, Kadowaki T, Lazar MA, O'Rahilly S, Palmer CN, Plutzky J, Reddy JK, Spiegelman BM, Staels B, Wahli W. International Union of Pharmacology. LXI. Peroxisome proliferatoractivated receptors. Pharmacol Rev 2006; 58:726 - 741
- Schrader M, Fahimi HD. Growth and division of peroxisomes. Int Rev Cytol 2006; 255:237 - 290
- Palma JM, Garrido M, Rodríguez García MI, del Río LA. Peroxisome proliferation and oxidative stress mediated by activated oxygen species in plant peroxisomes. Arch Biochem Biophys 1991; 287:68 - 74
- Lopez Huertas E, Charlton WL, Johnson B, Graham IA, Baker A. Stress induces peroxisome biogenesis genes. EMBO J 2000; 19:6770 - 6777
- Castillo MC, Sandalio LM, Del Río LA, León J. Peroxisome proliferation, wound-activated responses and expression of peroxisome-associated genes are cross-regulated but uncoupled in Arabidopsis thaliana. Plant Cell Environ 2008; 34:492 - 505
- Lingard MJ, Trelease RN. Five Arabidopsis peroxin 11 homologs individually promote peroxisome elongation, duplication or aggregation. J Cell Sci 2006; 119:1961 - 1972
- Orth T, Reumann S, Zhang X, Fan J, Wenzel D, Quan S, Hu J. The PEROXIN11 protein family controls peroxisome proliferation in Arabidopsis. Plant Cell 2007; 19:333 - 350
- Mano S, Nakamori C, Kondo M, Hayashi M, Nishimura M. An Arabidopsis dynamin-related protein, DRP3A, controls both peroxisomal and mitochondrial division. Plant J 2004; 38:487 - 498
- Nila AG, Sandalio LM, López MG, Gómez M, del Rio LA, Gómez-Lim MA. Expression of a peroxisome proliferator-activated receptor gene (xPPARalpha) from Xenopus laevis in tobacco (Nicotiana tabacum) plants. Planta 2006; 224:569 - 581
- Desai M, Hu J. Light induces peroxisome proliferation in Arabidopsis seedlings through the photoreceptor phytochrome A, the transcription factor HY5 HOMOLOG, and the peroxisomal protein PEROXIN11b. Plant Physiol 2008; 146:1117 - 1127