Abstract
EIN3 is a key transcription factor in the ethylene signaling pathway in Arabidopsis and its action is controlled by SCFEBF1/2 complex-mediated ubiquitination and subsequent 26S proteasome-dependent degradation. Our recent study revealed that ethylene signaling is regulated by a negative feedback system involving the direct activation of the EBF2 promoter by EIN3. This negative feedback loop likely modulates both the magnitude of the response to ethylene and the recovery after its withdrawal. The sequence downstream of the EBF2 stop codon was also found to play a role in determining the EBF2 expression levels independently of ethylene signaling. The reduction in EBF2 expression via the function of this sequence was suggested to be important for coupling the EBF2 expression levels to growth control. Two different mechanisms thus appear to be involved in determining ethylene sensitivity in Arabidopsis.
Addendum to: Konishi M, Yanagisawa S. Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. Plant J 2008; In press.
Ethylene Sensitivity is Controlled by Feedback Regulation Involving the Direct Activation of the EBF2 gene by EIN3
Ethylene is a gaseous plant hormone involved in a variety of processes including growth control, fruit ripening, disease resistance and senescence. The mechanisms underlying ethylene perception and the subsequent transduction of this signal have been investigated previously by both genetic and biochemical analyses.Citation1,Citation2 The accumulation of the EIN3 (ETHYLENE INSENSITIVE 3) transcription factor has been identified as a key step in ethylene signaling.Citation3–Citation6 The EIN3 protein levels are low in the absence of ethylene due to their constitutively active ubiquitination and subsequent degradation by the 26S proteasome. EIN3 is stabilized and begins to accumulate upon the transmission of ethylene signals leading to activation of its target genes, including the ERF1 transcription factor gene. The ubiquitination of EIN3 is mediated by the E3 complex, SCFEBF1/2, containing either one of the two closely related F-box proteins EBF1 and EBF2. Given their role as E3 subunits that selectively recognize EIN3,Citation4–Citation6 these F-box proteins are thus important components in the ethylene signaling pathway.
Our recent analyses of the EBF gene expression control have revealed that direct activation of the EBF2 promoter by EIN3 induces a feedback regulation in ethylene signaling.Citation7 The regulatory loop is illustrated in , on the basis of our recent findings, including the in vitro binding of EIN3 to a 5′-TACAT-3′ site in the EBF2 promoter (−230 to −226 relative to the translation start site) and also the transactivation of the EBF2 promoter by EIN3 in vivo. The significance for the interaction of EIN3 with the EBF2 promoter is demonstrated by the fact that a mutation in the EIN3-binding site prevents its transactivation in protoplasts and directs the ethylene-insensitive expression of a GUS reporter gene in transgenic Arabidopsis. This contrasts with the wild-type EBF2 promoter. Hence, EIN3 can activate the transcription of downstream genes in the ethylene signaling pathway and function at the same time as a component of the feedback regulation of this pathway. The maximal ethylene signal is likely to be transmitted to downstream components during a lag period prior to its downregulation by this negative feedback.
The physiological relevance of transactivation of the EBF2 promoter by EIN3 has been clarified by complementation analyses of the ebf2 mutant. In contrast to the wild-type EBF2 promoter, a mutant EBF2 promoter cannot confer ethylene-inducible expression of the gene. Hence, ebf2 mutants transformed with an exogenous EBF2 gene under the control of a mutant EBF2 promoter and non-transformed ebf2 mutants show the same ethylene hyper-responsiveness (mut-EBF2 3′ in ). This indicates that the activation of EBF2 transcription by EIN3 is indispensable for the function of this gene in vivo and that the regulatory feedback loop is required to maintain a normal ethylene-responsiveness, at least under persistent exposure to ethylene. As a recent kinetic study has indicated that the ebf2 mutant also exhibits a delay in recovery after the removal of ethylene,Citation8 the negative feedback control of ethylene signaling presumably also modulates this process.
Our findings suggest that the direct activation of EBF2 expression by EIN3 reflects a physiological role for this gene that is separate from the role for EBF1. Indeed, EBF1 and EBF2 similarly promote EIN3 degradation,Citation7 but the corresponding ebf1 and ebf2 mutants show different sensitivities to exogenously supplied ethylene.Citation4–Citation6,Citation8
Requirement for a Constitutive Reduction in EBF2 Expression for Precise Ethylene Signaling
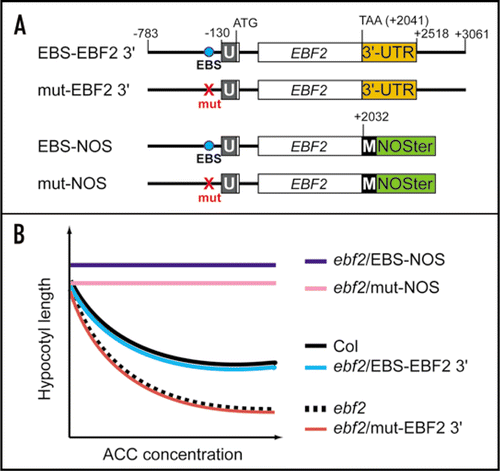
In addition to the EIN3-binding site, the sequence downstream of the open reading frame (ORF) of the EBF2 gene was also found to play a role in regulating its expression levels. When the nopaline synthase terminator (NOSter) was substituted for this region of the gene, the levels of EBF2 mRNA were found to be higher in the corresponding transgenic Arabidopsis. As a consequence, and presumably due to the reduced levels of EIN3 protein, the ebf2 mutant transformed with this EBF2-NOSter construct displayed complete ethylene insensitivityCitation7 (EBS-NOS in ). This suggested that sequence elements downstream of the EBF2 ORF play a critical role in regulating accumulation of EBF2 mRNA and also the ethylene sensitivity levels.
In a further experiment, we expressed the EBF2-NOSter chimeric gene under the control of a mutated EBF2 promoter (mut-NOS in ). The resulting transformants were found to be resistant to exogenously supplied ethylene. The hypocotyl lengths of these transformants were also longer than wild-type Columbia and shorter than ebf2 mutant plants harboring the EBF2-NOSter chimeric gene under wild-type EBF2 promoter control (compare EBS-NOS and mut-NOS in ). As the hypocotyl length is an indicator of the ethylene-dependent triple response in Arabidopsis, our findings suggest that the EIN3 transcription factor and the sequence downstream of the EBF2 ORF independently control EBF2 expression.
We do not yet have data that elucidates the mechanism underlying the negative effects of the EBF2 downstream sequence. Because both the 3′-UTR and a 543 bp region of the 3′-flanking sequence were used in our investigation of the role of this sequence,Citation7 we cannot currently exclude the possibility that a negative cis-element in this region may repress transcription. However, it is more likely that the 3′-UTR sequence of EBF2 mRNA renders the transcript unstable. Indeed, the half-life of EBF2 mRNA has been shown to be short (approximately 42 to 58 min).Citation9,Citation10 Furthermore, recent studies have reported that mutations in a 5′→3′ exoribonuclease, EIN5/XRN4, lead to overaccumulation of the EBF1 and EBF2 transcripts and cause an ethylene-insensitive phenotype.Citation11–Citation13 Because 5′→3′ exoribonuclease is involved in the degradation of 5′-decapped mRNA and also mRNA fragments that have arisen from internal cleavage events such as miRNA-mediated cleavage,Citation13–Citation15 we speculate that EIN5/XRN4 might play a role in the 3′-UTR-dependent rapid degradation of EBF1 and EBF2 transcripts. An investigation of the relationship between EIN5/XRN4 and the effects caused by a removal of the regulatory sequence downstream of the EBF2 ORF might provide new insights into ethylene signaling.
Although the molecular mechanisms that upregulate EBF2 expression following the removal of the regulatory region downstream of the EBF2 ORF are largely unknown, the phenotype resulting from this manipulation is noteworthy.Citation7 Although the basal EBF2 mRNA levels in transgenic Arabidopsis harboring the EBF2-NOSter construct were found to be already higher than that in the wild-type Columbia in the absence of exogenously supplied ethylene, these levels were further increased in response to ethylene.Citation7 This indicates that the ethylene-promoted accumulation of EIN3 occurs normally in these transgenic lines. However, these transgenic seedlings showed no ethylene-induced shortening of hypocotyls (). This suggests that the sequence downstream of the EBF2 ORF plays an essential role in both maintaining the EBF2 transcript level within an appropriate range, independently of ethylene signaling, and also in the coupling of ethylene-responsive gene expression to growth control.
Conclusions
The expression of EBF2 is regulated by two separate mechanisms. One is an ethylene-induced and EIN3-mediated system that produces negative feedback regulation of ethylene signaling. Another is ethylene signaling-independent and results in a constitutively lower level of EBF2 expression. We contend therefore that a precise ethylene response in Arabidopsis relies on both ethylene signaling-specific and -independent mechanisms.
Figures and Tables
Figure 1 A model of the feedback regulation loop mediated by EIN3 and EBF2 in ethylene signaling. EIN3 is ubiquitinated by the SCFEBF complex and then degraded by the 26S proteasome. Ethylene represses this degradation pathway and the resulting accumulation of EIN3 activates the transcription of target genes. Simultaneously, EIN3 binds and activates the EBF2 promoter. EBF2 is recruited into an SCF complex (SCFEBF2) that promotes the ubiquitination and 26S proteasome-mediated degradation of EIN3.
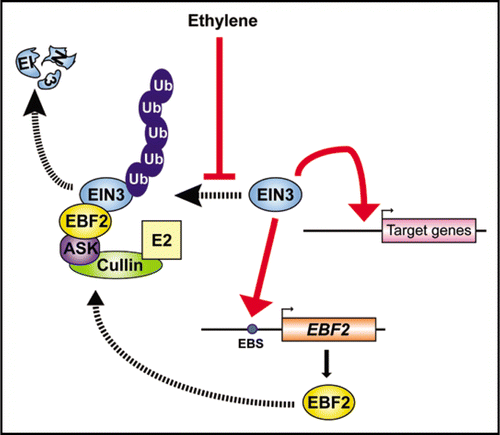
Figure 2 Requirement of two different regions of EBF2 for its precise expression. (A) Schematic representation of constructs used to transform the ebf2 mutant. These constructs contained either a DNA fragment corresponding to the region from −783 to +3061 of the EBF2 gene (relative to the translation initiation Met codon) or from −783 to +2032 fused to the NOS terminator (NOSter). The EIN3-binding site (EBS) and its mutated version are indicated by “EBS” and “mut”, respectively. The 5′ untranslated region and the MYC-tag are indicated by U and M, respectively. (B) Schematic representation of the ethylene sensitivity profiles of wild-type Columbia (Col), ebf2 and ebf2 transformed with the constructs listed in (A). Because ethylene induces a triple response that includes a shortening of the hypocotyls, the ethylene response is evaluated by measuring the hypocotyl lengths of etiolated seedlings that were grown in the presence of increased concentrations of 1-amino-1-cyclopropanecarboxylic acid (ACC), an immediate precursor of ethylene. Mutation of the EBS abolishes the complementation of the ethylene hyper-responsive phenotype of ebf2 (ebf2/mut EBF2 3′). The use of NOSter resulted in an ethylene-insensitive phenotype (ebf2/EBS-NOS and ebf2/mut-NOS). Note that the insertion of a MYC-tag at the 3′ end of the EBF2 ORF did not affect the ethylene sensitivity.Citation7
Acknowledgements
This work was supported by CREST, JST.
Addendum to:
References
- Bleecker AB, Kende H. Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol 2000; 16:1 - 18
- Guo H, Ecker JR. The ethylene signaling pathway: new insights. Curr Opin Plant Biol 2004; 7:40 - 49
- Yanagisawa S, Yoo SD, Sheen J. Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 2003; 425:521 - 525
- Guo H, Ecker JR. Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell 2003; 115:667 - 677
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 2003; 115:679 - 689
- Gagne JM, Smalle J, Gingerich DJ, Walker JM, Yoo SD, Yanagisawa S, Vierstra RD. Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc Natl Acad Sci USA 2004; 101:6803 - 6808
- Konishi M, Yanagisawa S. Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. Plant J 2008; In press
- Binder BM, Walker JM, Gagne JM, Emborg TJ, Hemmann G, Bleecker AB, Vierstra RD. The Arabidopsis EIN3 binding F-box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell 2007; 19:509 - 523
- Gutiérrez RA, Ewing RM, Cherry JM, Green PJ. Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc Natl Acad Sci USA 2002; 99:11513 - 11518
- Narsai R, Howell KA, Millar AH, O'Toole N, Small I, Whelan J. Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 2007; 19:3418 - 3436
- Olmedo G, Guo H, Gregory BD, Nourizadeh SD, Aguilar-Henonin L, Li H, An F, Guzman P, Ecker JR. ETHYLENE-INSENSITIVE5 encodes a 5′→3′ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc Natl Acad Sci USA 2006; 103:13286 - 13293
- Potuschak T, Vansiri A, Binder BM, Lechner E, Vierstra RD, Genschik P. The exoribonuclease XRN4 is a component of the ethylene response pathway in Arabidopsis. Plant Cell 2006; 18:3047 - 3057
- Souret FF, Kastenmayer JP, Green PJ. XRN4 degrade mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell 2004; 15:173 - 183
- Kastenmayer JP, Green PJ. Novel features of the XRN-family in Arabidopsis: Evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc Natl Acad Sci USA 2000; 97:13985 - 13990
- Amrani N, Sachs MS, Jacobson A. Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol 2006; 7:415 - 425