Abstract
High salinity is an important factor limiting agriculture as major crops are salt sensitive. Understanding salt stress signalling is key to producing salt tolerant crops. The small ubiquitin-like modifier (SUMO) is a crucial regulator of signalling proteins in eukaryotes. Attachment of SUMO onto substrates is reversible and SUMO-proteases which specifically cleave the SUMO-substrate linkages play a vital regulatory role during SUMOylation. We have identified two SUMO proteases OTS1 and OTS2 that act redundantly to regulate salt stress responses in Arabidopsis. ots1 ots2 double mutants show extreme sensitivity to salt. However during non-salt conditions, ots1 ots2 double mutants are phenotypically similar to wild-type plants in terms of growth and development. Overexpressing SUMO1 in the ots1 ots2 double mutants severally diminishes plant size as quantified by rosette diameter even under non-stressed conditions. This reduction in plant growth is reminiscent of ots1 ots2 double mutants under salt stress. Our data indicates that overSUMOylation of target proteins can have severe effects on plant growth and that SUMO proteases like OTS1/2 are key to maintaining cellular balance of SUMOylation. We propose that upon environmental stress the hyperSUMOylation of key target proteins act to retard growth to survive stress periods.
Environmental stress caused by high salinity is a major factor limiting plant growth and productivity. It is increasingly evident that developing salt tolerant high-yield crops will be a key objective if global food production is to be maintained.
Plants tolerate salt stress conditions by a variety of biochemical and physiological mechanisms. In Arabidopsis, this includes the restoration of salt balance in the cell, increased efficiency of water use, induction of ROS detoxifying agents, altered gene expression and a reduction of growth rate.Citation2 However, the molecular mechanisms allowing the plant to mount these responses to adapt their growth and development in adverse conditions is poorly understood.
Post-translational modification of proteins plays a critical role in most cellular signalling processes. In eukaryotes, an important form of such modifications is the attachment of a small polypeptide tag to a target protein and Ubiquitin (Ub) is the best understood tag. In recent years one such class of Ub-related tags called small ubiquitin-like modifiers (SUMO) has emerged as a very influential molecular regulator.Citation3 SUMO maturation and attachment on to substrates is similar to the ubiquitination process with its own set of analogous E1, E2 and E3 enzymes involved in activation, conjugation and ligation respectively. Many important regulatory proteins modified by SUMO have been described in yeast, mammals and Drosophila.Citation4
The importance of SUMOylation in plants is just beginning to be discovered. Different SUMO isoforms are present in Arabidopsis although only the two nearly identical AtSUMO1 and AtSUMO2 isoforms are conjugated onto target proteins upon various types of stress.Citation5 Mutants that fail to promote SUMO 1/2 attachment onto target proteins display a number of phenotypes including reduced growth, altered response to phosphate starvation, deregulated salicylic acid (SA) dependant signalling upon pathogen infection, reduced basal thermotolerance and impaired drought and freezing tolerance.Citation6–Citation8 Attachment of SUMO onto substrates is also reversible and the same key enzymes involved in generating mature SUMO—the SUMO-proteases—also act as isopeptidases to specifically cleave the SUMO-substrate linkages to recycle free SUMO. A considerable fraction of protein substrates are normally SUMOylated at a given time, clearly implying that SUMO-proteases play a vital regulatory role during SUMOylation.Citation9
We have identified two SUMO proteases, OVERLY TOLERANT TO SALT1 (OTS1) and OTS2, which are localised in the nucleus and act redundantly to control salt stress responses in Arabidopsis. ots1 ots2 double knockout mutants show extreme sensitivity to salt. We demonstrated for the first time that salt stress can induce a dose-dependant accumulation of SUMO1/2-conjugated proteins in Arabidopsis.Citation9 ots1 ots2 double mutants show a further dramatic increase in SUMO1/2-conjugated proteins in response to salt stress while transgenic lines overexpressing OTS1, but not a protease deficient allele of OTS1, have increased salt tolerance and a concomitant reduction in the levels of SUMOylated proteins.Citation9 Our data demonstrate that OTS1/2 deSUMOylated target proteins are vital components of a plant's armoury against salt stress.
In Arabidopsis AtSUMO1/2 SUMOylation increases upon different types of stress including heat, cold and drought.Citation10,Citation11 Our data demonstrate a role for SUMOylation also in salt stress. Abundance of AtSUMO1/2 conjugates increased in a salt concentration dependent manner and correlated with inhibition of Arabidopsis seedling growth. Moreover, upon salt, the levels of free AtSUMO1/2 increased strongly in wild-type plants but to a much reduced extent in ots1 ots2 double mutants. This could be partly due to an overall reduced cellular deconjugation activity to recycle AtSUMO1/2 from their target proteins in ots1 ots2 mutants.
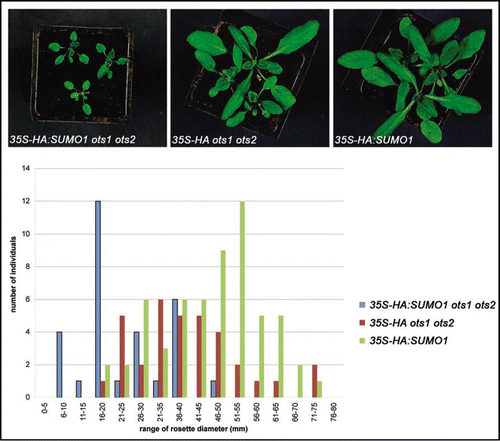
Salt-induced AtSUMO1/2 conjugation is directly linked to reduced growth. However, ots1 ots2 double mutants accumulated AtSUMO1/2-conjugates even under non-stress conditions but this did not produce any obvious growth inhibition. This data suggests that in ots1 ots2 double mutants the accumulation of these particular set of AtSUMO1/2 conjugates is largely dispensable for growth and survival under non-stress conditions. Alternatively, the levels of SUMO1/2 freely available for conjugation on to these targets might be rate limiting in ots1 ots2 implying that overexpression of SUMO1/2 may result in a significant reduction of growth even in normal conditions. To test these possibilities we have overexpressed HA-tagged SUMO1 in the wild-type and ots1 ots2 double mutant backgrounds and analysed several independent T1 transgenic plants. Overexpression of HA:SUMO1 through the 35S promoter (35S-HA:SUMO1) in the ots1 ots2 double mutant background resulted in a marked decrease in size compared to wild-type plants transformed with the same construct and this was quantified by measuring rosette diameter of individual plants (). Furthermore, no significant difference in rosette diameter was found between wildtype plants overexpressing HA:SUMO1 or ots1 ots2 plants expressing just the HA-tag (35S-HA). Our data suggests that OTS1/2 SUMO proteases are required even under unstressed conditions to maintain key target proteins in the de-SUMOylated state for proper plant growth and development. Presumably in the ots1 ots2 double mutant background a subset of target proteins that govern plant development are hyperSUMOylated compared to wild type but the overall level of the SUMOylated pool of these target proteins may not be large enough to have a physiological effect under non stressed conditions. However, overexpression of HA-SUMO1 further increases the SUMOylated pool of these target proteins. This greater shift in the balance between non-SUMOylated and SUMOylated forms in the 35S-HA:SUMO1 ots1 ots2 transgenics leads to impaired development and hence reduced plant size. It can be argued that the same shift in the SUMOylation status occurs in ots1/ots2 double mutant plants during salt stress resulting in salt sensitivity observed by reduced root and shoot growth.Citation9
Overall our data indicate that the target proteins of OTS1/2 not only control salt tolerance mechanisms but also form a link between salt stress and plant development. Furthermore, these 35S-HA:SUMO1 ots1 ots2 transgenic plants represent a useful tool to purify HA tag-SUMOylated proteins that are enriched in the ots1 ots2 double mutant background. The identification of OTS1/2 dependant SUMO-conjugate targets will undoubtedly provide valuable new information on not only salt tolerance mechanisms but also how plants perceive abiotic stress to modulate their development.
Figures and Tables
Figure 1 Arabidopsis wild-type (Columbia) or ots1-1 ots2-1 double mutantsCitation9 were transformed with pEarleyGate 201 vectors carrying N-terminal HA-tagged Arabidopsis SUMO1 with the floral dipping method.Citation12 T1 seeds were sown on Agar/MS plates containing BASTA (20 mg/L glufosinate ammonium) and after ten days BASTA resistant seedlings were transferred to soil under a 16 h of light/8 h of dark photoperiod at 22°C with a light intensity of 90 µmol·m−2·s−1. Two weeks later, T1 plants were photographed (upper) and the rosette diameter of each plant was measured and compared (lower).
Addendum to:
References
- Nelson DE, Shen B, Bohnert HJ. Setlow JK. Salinity tolerance: mechanisms, models and the metabolic engineering of complex traits. Genetic Engineering, Principles and Methods 1998; 20:New York Plenum Press 153 - 176
- Zhu JK. Plant salt tolerance. Trends Plant Sci 2001; 6:66 - 71
- Melchior F. SUMO—nonclassical ubiquitin. Annu Rev Cell Dev Biol 2000; 16:591 - 626
- Hay RT. SUMO-specific proteases: a twist in the tail. Trends Cell Biol 2007; 17:370 - 376
- Saracco SA, Miller MJ, Kurepa J, Vierstra RD. Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol 2007; 145:119 - 134
- Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, et al. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 2007; 49:79 - 90
- Miura K, Jin JB, Hasegawa PM. Sumoylation, a post-translational regulatory process in plants. Curr Opin Plant Biol 2007; 10:495 - 502
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, et al. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 2007; 19:1403 - 1414
- Conti L, Price G, O'Donnell E, Schwessinger B, Dominy P, Sadanandom A. Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 2008; 20:2894 - 2908
- Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, et al. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem 2003; 278:6862 - 6872
- Catala R, Ouyang J, Abreu IA, Hu Y, Seo H, Zhang X, et al. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 2007; 19:2952 - 2966
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 1998; 16:735 - 743