Abstract
The phytohormone abscisic acid (ABA), an important bioactive compound in plants, is implicated in several essential processes such as development and the abiotic stress response. Many components have been reported to have roles in these processes. Although 2C-type protein phosphatases (PP2C) and SNF1-related protein kinases (SnRK2) are known to be important signal mediators, the molecular mechanisms by which these components regulate the ABA signaling pathway have not been elucidated. Recent identification of soluble ABA receptors, PYR/PYL/RCAR, has provided a major breakthrough in understanding the signaling mechanisms of ABA and revealed the importance of PP2Cs. In addition, the physical, biochemical, and physiological connections between PP2C and SnRK2 have been clearly demonstrated. Taken together, the molecular basis of the major ABA signaling pathway has been established, from perception to gene expression. In this addendum, we discuss this emerging ABA signaling pathway, which has a conventional protein phosphorylation/dephosphorylation regulatory circuit and consider its physiological and functional relevance.
Introduction
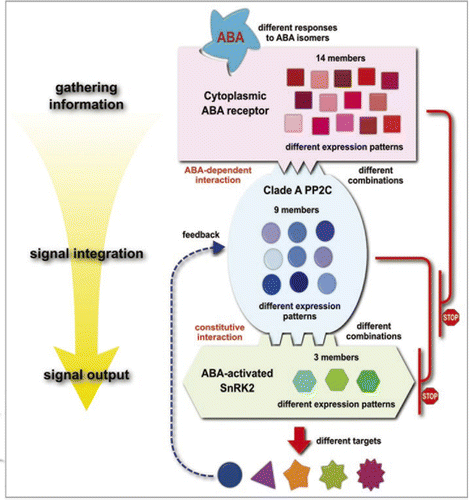
The phytohormone abscisic acid (ABA) has broad physiological functions, not only in the abiotic stress response, but also in developmental processes and the biotic stress response. Its complex physiological activity and huge amount and variety of putative ABA signaling factors suggest that the ABA signaling pathway is a complex network.Citation1,Citation2 However, the emerging picture of the ABA signaling pathway has changed our perception drastically. Recent breakthrough studies have revealed that PYR/PYL/RCAR family proteins function as soluble ABA receptors. These ABA receptors regulate clade A PP2Cs in an ABA-dependent manner. Citation3,Citation4 We very recently showed that these PP2Cs interact with and inactivate ABA-activated SNF1-related kinase2 (SnRK2) via dephosphorylation and that this PP2C action is inhibited by PYR1 in the presence of ABA.Citation5 These studies have established one of the major ABA signaling pathways consisting of just four factors, PYR/PYL/RCAR ABA receptors—clade A PP2Cs—ABA activated SnRK2s—targets of kinase (e.g., bZIP)(). This pathway is as simple as the signaling systems of other plant hormones such as auxin, gibberellin, or jasmonic acid.Citation6 In this addendum, we will review the regulatory steps of the ABA signaling pathway and discuss the relevance of using a phosphorylation/dephosphorylation modification system in the signal transduction pathway of this hormone.
Soluble ABA Receptors
PYR/PYL/RCAR proteins have been identified using a chemical genetic research strategyCitation4 or biochemical approaches.Citation3 Fine biochemical studies showed that those proteins bind ABA efficiently. These proteins, however, have no known functional domains but they bind clade A PP2Cs such as ABI1, ABI2, HAB1, HAB2, AHG1 and AHG3/AtPP2CA. Arabidopsis has a total of 14 PYR/PYL/RCAR-related proteins, many of which bind to several PP2Cs in yeast two-hybrid experiments. Moreover, PYR1, RCAR1, PYL5 and RCAR3 inhibit PP2C activity in an ABA-dependent manner. Therefore, PYR/PYL/RCAR receptors are presumed to transmit the ABA signal through clade A PP2Cs.Citation3,Citation4,Citation7,Citation8
Clade A PP2Cs are known as major negative regulators of ABA signaling.Citation9–Citation11 Originally, this type of PP2C was identified by a genetic screen in which the ABA insensitive1-1 (abi1-1) and abi2-1 mutation were isolated.Citation12,Citation13 These mutations have a single amino acid substitution at the same corresponding site. The abi1-1 mutant PP2C has strongly reduced phosphatase activity in the conventional phosphatase assay using artificial substrates such as casein, which does not explain the phenotype. Strikingly, abi1-1 or abi2-1 mutant PP2Cs do not bind ABA receptors.Citation3,Citation4 This property of the abi1-1-type mutant PP2C may explain the ABAinsensitive phenotype of the abi1-1 or abi2-1 mutants. However, the low PP2C activity of abi1-1 PP2C cannot completely explain the phenotype. The molecular basis for the action of abi1-1-type mutation has remained unclear.
Hence, an important question arises: what is the target of PP2C? Several proteins have been reported as target candidates of PP2C due to their ability to interact with PP2Cs.Citation2 Among these reports, a demonstration of an interaction between ABI1 and SRK2E/OST1, an SnRK2, is intriguing because several SnRK2s are known to have important roles in the ABA response.Citation14 Furthermore, the activation of SnRK2 is impaired in the abi1-1 mutant, suggesting the upstream regulation of PP2C. However, the physiological relevance of this interaction has not been elucidated. Park et al. proposed a model in which SnRK2 is regulated by a PYR/PYL/RCAR-PP2C complex based on their data showing a defect in the activation of SnRK2s in multiple loss-of-ABA receptor mutants. A direct demonstration of physical and biochemical interactions between PP2Cs and SnRK2s has not been performed.Citation4
Physical and Biochemical Interaction between PP2C and SnRK2
In our recent publication,Citation5 we demonstrated that clade A PP2Cs consistently bind ABA-activated SnRK2s with varied preferences, but not ABA-independent SnRK2s in yeast two-hybrid analysis. In planta interactions were also found in several cases. Our data suggest that the interaction is constitutive and ABA-independent. More importantly, we showed that PP2C inactivates SnRK2 kinase activity through direct dephosphorylation. Using nanoLC/MS analysis, several Ser/Thr residues on the kinase activation loop were identified as relevant phosphorylation sites. Taken together, these data suggest that PP2C negatively regulates SnRK2 by dephosphorylation in the PP2C-SnRK2 complex.
Notably, we found that the abi1-1-type mutant PP2Cs, abi1-1 and ahg1G152D, behave as a wild-type PP2C for SnRK2.Citation5 The mutant PP2C bound SnRK2 and inactivated the kinase activities. Unexpectedly, the mutant PP2Cs dephosphorylated SnRK2s with comparable activity to the wild-type PP2C, despite the fact that these mutant PP2Cs had very low PP2C activity against artificial substrates such as a phosphopeptide or phosphorylated casein. These data strongly suggest that the abi1-1-type mutation does not significantly affect phosphatase activity of clade A PP2Cs against ABA-activated SnRK2s and imply that these SnRK2s are native substrates for these PP2Cs.
Connecting Nodes from Perception to Gene Regulation
Upon identification of soluble ABA receptors, in vitro reconstitution of the receptor-PP2C-SnRK2 complex was attempted.Citation5 In the presence of recombinant ABI1 and PYR1, the ABI1-dependent negative regulation of SRK2E was canceled out in an ABA-dependent manner. In contrast, the abi1-1 mutant PP2C constitutively inactivated SRK2E even in the presence of ABA. As SnRK2 has been shown to phosphorylate and activate bZIP transcription factors ABF/AREBs,Citation15,Citation16 a major ABA signaling cascade from ABA perception to gene regulation has been established: ABA → PYR/PYL/RCAR → PP2C → SnRK2 → bZIP → gene expression. Given no visible ABA responses in the triple loss-of-function mutant of ABA-activated SnRK2s, this pathway could be the major ABA signaling pathway.
Discussion and Perspective
Finally, many genetic and biochemical studies have established a major ABA signaling pathway, which consists of four components, PYR/PYL/RCARPP2C-SnRK2—targets of SnRK2 (e.g., transcription factor). Given the picture of this new ABA signaling pathway, several questions arise. Arabidopsis has 14 receptors, nine clade A PP2Cs, and three ABA-activated SnRK2s. Why are so many redundant components involved in ABA signaling and why are signals perceived by multiple receptors converged to only three components at SnRK2s? Presumably, these receptors have different characteristics so as to monitor a broad range of ABA states, that is, several orders of ABA concentration or different isomer compositions at the first step of the ABA signaling cascade. Indeed, receptors seem to have different selectivity to isomers of ABA.Citation4,Citation8 Their different expression patterns are also consistent with this idea.Citation4 At the second step, the ABA signal is converted through the reduction of PP2C activity. At this point, signals of 14 receptors are reduced to nine PP2Cs. Some selectivity can occur between receptors and PP2Cs. For example, PYL5 binds ABI1, HAB1 and ABI2 but not AHG3.Citation7 The third step is the regulation of SnRK2 by PP2C. The two-hybrid test in yeast cells shows some preferences in the interaction between these two proteins.Citation5 Indeed, the subcellular localization of PP2C differs among clade A PP2Cs. For example, ABI1 and ABI2 are dispersed throughout the whole cell, while AHG1 and AHG3 localize mainly to the nucleus. These preferences in the interactions among receptors, PP2Cs, and SnRK2s might reflect their different physiological roles in the ABA response. Many proteins other than SnRK2s have been reported as substrates for PP2Cs. The possibility exists that not only SnRK2s, but also these other proteins, are the target of ABA responses, especially in some specific tissues (e.g., reproductive tissues or stomata) or developmental stages (seed development, maturations or germination). For such specific ABA responses, direct regulation of these targets by PP2Cs might be required. Collectively, the emerging picture indicates that the major ABA signaling pathway has both flexibility and robustness.
The signaling mechanisms of the major phytohormones, auxin, cytokinin, gibberellin (GA), ethylene, jasmonic acid (JA) and ABA, have now been elucidated.Citation6 This ABA signaling pathway resembles the signaling mechanisms of auxin, GA and JA, in which receptors promote derepression of transcription factors in a hormone-dependent manner. So far, the direct targets of these hormone receptors are not known, except for repressors for transcription factors. In the case of ABA, the receptor induces activation of protein kinase (SnRK2), which in turn activates transcription factors. Presumably, besides transcription factors, SnRK2 has other targets such as RNA-binding proteins and AtrbohF NADPH oxidase.Citation17,Citation18 Using SnRK2 as an effecter, the ABA signaling system can promptly evoke wider cellular responses. In addition, the direct target of the ABA receptor, PP2C, is thought to have substrates other than SnRK2s. Inhibition of PP2Cs may add another dimension of response. Therefore, one could postulate that ABA chooses kinases/phosphatases as main effectors in order to drive complex cellular responses.
Another unique factor in the ABA signaling pathway could be the desensitization. In the other three plant hormone systems, interaction with a receptor promotes degradation of target proteins, which in turn releases and activates the transcription factors. These processes are irreversible, and de novo protein synthesis is required for desensitization. Interestingly, as described above, ABA presumably evokes a reversible process. This can be explained by the broadness of the effect of ABA on cellular processes. Desensitization of such a signal should be executed quickly. Alternatively, a reversible signaling mechanism could be applied to fulfill the physiological requirement under adverse conditions in which ABA plays important roles. Because cellular processes are restricted under such conditions, a reversible process is suitable for the desensitization.
Many questions, however, remain on ABA signaling. For example, the molecular mechanism of SnRK2 activation and mechanisms of other ABA signaling pathways have not been resolved. Further analysis is required to fully understand the ABA response, which will help produce a clearer picture of the stress response mechanisms of plants.
Figures and Tables
Figure 1 Schematic representation of the major ABA signaling pathway. Under normal conditions, clade A PP2C constantly interacts with ABA-activated SnRK2 and inactivates its kinase activity by dephosphorylation. When the soluble ABA receptor recognizes the ABA molecule, it binds PP2C and inhibits its phosphatase activity. Then SnRK2 is activated and phosphorylates its targets, and the ABA signal is consequently turned on. In this ABA signaling pathway, the signal is gathered by 14 ABA receptors and transduced and integrated through nine PP2Cs and three SnRK2s. Presumably, in these steps, the signal information is processed by different interactions between components for correct cellular responses.
Acknowledgements
We thank Dr. Shinozaki for fruitful discussions. This work was partly supported by Grants-in-Aid for Scientific Research (C) to T.H. and Grants-in-Aid for Scientific Research for Young Scientists (B) to T.U. from MEXT.
Addendum to:
References
- Finkelstein R, Gibson S. ABA and sugar interactions regulating development: cross-talk or voices in a crowd?. Curr Opin Plant Biol 2002; 5:26 - 32
- Hirayama T, Shinozaki K. Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci 2007; 12:343 - 351
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009; 324:1064 - 1068
- Park S, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009; 324:1068 - 1071
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, et al. Type 2C protein phosphatases directly regulate abscisic acidactivated protein kinases in Arabidopsis. Proc Natl Acad Sci USA 2009; 106:17588 - 17593
- Santner A, Estelle M. Recent advances and emerging trends in plant hormone signalling. Nature 2009; 459:1071 - 1078
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, et al. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J 2009; in press
- Szostkiewicz I, Richter K, Kepka M, Demmel S, Ma Y, Korte A, et al. Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J 2009; in press
- Gosti F, Beaudoin N, Serizet C, Webb A, Vartanian N, Giraudat J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 1999; 11:1897 - 1910
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, et al. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol 2006; 140:115 - 126
- Kuhn J, Boisson-Dernier A, Dizon M, Maktabi M, Schroeder J. The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol 2006; 140:127 - 139
- Leung J, Bouvier-Durand M, Morris P, Guerrier D, Chefdor F, Giraudat J. Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 1994; 264:1448 - 1452
- Leung J, Merlot S, Giraudat J. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 1997; 9:759 - 771
- Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem 2006; 281:5310 - 5318
- Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, et al. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J 2005; 44:939 - 949
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, et al. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA 2006; 103:1988 - 1993
- Li J, Kinoshita T, Pandey S, Ng C, Gygi S, Shimazaki K, et al. Modulation of an RNA-binding protein by abscisic-acid-activated protein kinase. Nature 2002; 418:793 - 797
- Sirichandra C, Gu D, Hu H, Davanture M, Lee S, Djaoui M, et al. Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett 2009; 583:2982 - 2986