Abstract
We recently established a proteome methodology for Arabidopsis leaf peroxisomes and identified more than 90 putative novel proteins of the organelle. These proteins included glutathione reductase isoform 1 (GR1), a major enzyme of the antioxidative defense system that was previously reported to be cytosolic. In this follow-up study, we validated the proteome data by analyzing the in vivo subcellular targeting of GR1 and the function of its C-terminal tripeptide, TNL>, as a putative novel peroxisome targeting signal type 1 (PTS1). The full-length protein was targeted to peroxisomes in onion epidermal cells when fused N-terminally with the reporter protein. The efficiency of peroxisome targeting, however, was weak upon expression from a strong promoter, consistent with the idea that the enzyme is dually targeted to peroxisomes and the cytosol in vivo. The reporter protein that was extended C-terminally by 10 amino acid residues of GR1 was directed to peroxisomes, characterizing TNL> as a novel PTS1. The data thus identify plant peroxisomal GR at the molecular level in the first plant species and complete the plant peroxisomal ascorbateglutathione cycle. Moreover, GR1 is the first plant protein that is dually targeted to peroxisomes and the cytosol. The evolutionary origin and regulatory mechanisms of dual targeting are discussed.
Massive amounts of hydrogen peroxide (H2O2) are produced during photosynthesis in peroxisomes by glycolate oxidase activity as part of the photorespiratory cycle.Citation1 Next to catalase, the ascorbate-glutathione cycle is the secondary scavenging system for H2O2 detoxification.Citation2–Citation4 The cycle comprises four enzymes, ascorbate peroxidase (APX), monodehydroascorbate reductase (MDAR), dehydroascorbate reductase (DHAR) and NADPH-dependent glutathione reductase (GR). GR plays a major physiological role in maintaining and regenerating reduced glutathione in response to biotic and abiotic stresses in plants.Citation5 Jiminez et al. (1997) provided biochemical evidence for the presence of the antioxidants ascorbate and glutathione and the enzymes of the ascorbate-glutathione cycle in pea peroxisomes.Citation6–Citation8 While Arabidopsis APX3, MDAR1 and MDAR4 have been characterized as peroxisomal isoforms,Citation9–Citation11 the molecular identity of plant peroxisomal GR and DHAR have not been determined in any plant species to date.Citation5 Arabidopsis encodes two GR and five DHAR isoforms that are either shown to be or predicted to be cytosolic, mitochondrial or plastidic.Citation12 We recently identified specific isoforms of GR (GR1, At3g24170) and DHAR (DHAR1, At1g19570) as being peroxisome-associated by proteome analysis of Arabidopsis leaf peroxisomes.Citation13,Citation14 Both isoforms were previously reported to be or predicted to be cytosolic.Citation15
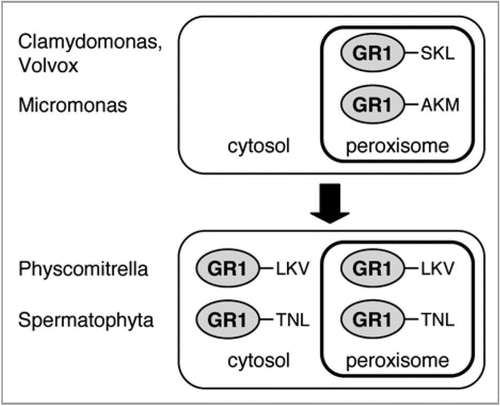
Arabidopsis GR1 terminates with TNL>, which is related to functional plant PTS1 tripeptides such as SNL> and ANL>.Citation16,Citation17 Threonine (T), however, has not yet been described as an allowed residue at position −3 of PTS1s in any plant peroxisomal protein.Citation16 Analysis of homologous plant proteins and expressed sequence tags (ESTs) shows that TNL> is generally highly conserved in putative plant GR1 orthologs (). A few other sequences terminate with related tripeptides, such TSL>, TTL>, NNL> and TKL>. Only a single EST (Picrorhiza kurrooa) carries the canonical PTS1, SKI> (). The data provide only weak additional support for peroxisome targeting of plant GR1 orthologs. However, GR homologs from green algae (chlorophyta) carry canonical PTS1 tripeptides, such as SKL> (Chlamydomonas, Volvox) and AKM> (Micromonas, , Suppl. ).
Arabidopsis GR1 is Targeted to Peroxisomes
To investigate peroxisomal targeting of GR1 by TNL> in vivo, we fused GR1 N-terminally with the reporter protein enhanced yellow fluorescent protein (EYFP) and expressed the construct transiently from the 35S cauliflower mosaic virus (CaMV) promoter in onion epidermal cells and tobacco protoplasts. In parallel, onions were routinely transformed with appropriate negative control plasmids, such as EYFP alone (), to verify the absence of endogenous bacteria that may phagocytose cytosolic GFP variants. Although they differed slightly in their active modes of movement, these GFP-labeled subcellular bacteria could, particularly in cases of weak targeting signals and efficiency, be misinterpreted as peroxisomes and cytosolic reporter protein fusions could be falsely identified as being peroxisomally targeted. The N-terminal full-length reporter protein fusion of GR1 (EYFP-GR1) was indeed targeted to small subcellular organelles (), but the efficiency of organelle targeting was weak, making characterization of the structures as peroxisomes difficult in double transformants. In a few cells that co-expressed a peroxisome marker (gMDH-CFP), EYFP-GR1 could conclusively shown to be peroxisomal. The data establish GR1 as a novel peroxisomal enzyme (). In tobacco protoplasts of leaf mesophyll cells, however, EYFP-GR1 remained exclusively cytosolic (), strengthening the idea that negative organelle targeting data may be related to the nature of the analytical in vivo system and require validation in an alternative plant expression system. Notably, the tobacco GR1 ortholog that has been assembled from overlapping ESTs also terminates with TNL> (, Suppl. ).
TNL> is a Novel Plant PTS1
To investigate whether AtGR1 is targeted to peroxisomes by TNL>, we fused the putative peroxisome targeting domain (PTD) comprising the C-terminal ten amino acid (aa) residues of GR1 to EYFP. The extended reporter protein was directed to small subcellular organelles that coincided with peroxisomes (). The peroxisome targeting efficiency ranged from weak to moderate. The same reporter construct also localized to peroxisomes in tobacco mesophyll protoplasts (). In both plant expression systems, the efficiency at which the C-terminal 10 aa residues of GR1 targeted EYFP to peroxisomes was considerably higher compared to full-length GR1. A possible explanation is that the full-length GR1 fusion protein is much larger than the C-terminal 10 aa residues. Due to the C-terminal location of the PTS1 sequence, incomplete transcription and translation products of the gene fusion could remain cytosolic. Alternatively, both EYFP constructs may differ in PTS1 accessibility.
The identification of TNL> as a functional PTS1 tripeptide is important for plant PTS1 protein prediction. First, seven additional Arabidopsis proteins terminating with TNL> emerge as candidate peroxisomal proteins (Suppl. Table 1). Second, the data establish T as a new allowed residue in plant PTS1 tripeptides. Six further tripeptides carrying T at pos. −3 (e.g., T[RK][LMI]>) are likely to represent additional functional PTS1 tripeptides. Third, C-terminal tripeptides that are found in a significant number of homologs of established plant PTS1 proteins are considered to be functional plant PTS1 tripeptides. Plant orthologs of GR1 carry a number of non-canonical C-terminal tripeptides, such as TSL> (), which are likely to emerge as additional plant PTS1 tripeptides in the near future.
Questions arise as to why the PTS1 TNL> has not been detected previously in plant homologs of PTS1 proteins,Citation16 and why it appears to be restricted to GR1 and possibly a few other proteins. First, the relatively weak targeting efficiency of EYFP-GR1 indicates that the PTS1 domain of GR1 is likely to be insufficient for quantitative peroxisomal targeting of high-abundance peroxisomal matrix proteins. Second, TNL> alone is predicted to be an extremely weak PTS1 tripeptide, requiring auxiliary targeting enhancing elements (e.g., K, P) upstream of the tripeptide (AHKPKPKTNL>) for moderate peroxisome targeting efficiency.Citation16,Citation18 Most single aa residue mutations, for instance, SNL> to TNL>, are thus likely to be insufficient to maintain peroxisome targeting if they are not paralleled by the conversion of upstream residues to those that enhance targeting. Such multiple mutations, however, are unlikely to occur simultaneously during protein evolution.
Dual Targeting of GR1 to Peroxisomes and the Cytosol
Together with identification of the peroxisomal isoform of DHAR (DHAR1, At1g19570,Citation14), all four members of the peroxisomal ascorbate-glutathione cycle have been identified in Arabidopsis at the molecular level. The pronounced conservation of TNL> in higher plant orthologs (, Suppl. ) strongly suggests that most plant GR1 orthologs, even though they are frequently annotated as cytosolic, are generally dually targeted to peroxisomes and the cytosol. Plant GR1 can thus regenerate reduced glutathione in the peroxisome matrix and substantially contribute to H2O2 detoxification. Moreover, peroxisomal GR1 provides the substrate for several other glutathione-dependent enzymes that have been recently discovered in plant peroxisomes.Citation13,Citation14,Citation19 Glutathione is thereby emerging as a major antioxidant in plant peroxisomes, protecting peroxisomal and cellular enzymes against a range of peroxides, xenobiotics and possibly heavy metals.Citation20
Dual or even multiple subcellular targeting of a single protein to several compartments emerges as a more prevalent phenomenon than previously assumed. Six Arabidopsis proteins are dually targeted to mitochondria and peroxisomes,Citation21,Citation22 and three proteins are dually targeted to plastids and peroxisomes (watermelon Hsp70;Citation23 soybean aspartate aminotransferase;Citation24 Arabidopsis 6-phosphogluconolactonaseCitation13). GR1 is thus the first plant protein that is dually targeted to peroxisomes and the cytosol. In the green lineage, most species bear two GR genes as a result of an early gene duplication event. The two genes cluster into a clade of chloroplastic/mitochondrial isoforms with N-terminal extensions and a clade of cytosolic/peroxisomal isoforms terminating with known or PTS1-related tripeptides (Suppl. ). PTS1 conservation analysis in cytosolic/peroxisomal isoforms allows the prediction as to whether the cytosolic or peroxisomal function of GR1 is the most fundamental and most evolutionarily ancient task. As revealed by phylogenetic analysis of full-length protein sequences and cDNAs assembled from ESTs, one of two GR homologs from green algae carries a strong canonical PTS1 tripeptide, such as SKL> (Chlamydomonas, Volvox) and AKM> (Micromonas). The data indicate that the peroxisome-targeting function of GR evolved soon after gene duplication in Viridiplantae and that these GR1 homologs are most likely exclusively peroxisomally targeted in chlorophyta (Suppl. , see also ). Dual targeting of GR1 to peroxisomes and the cytosol appears to have evolved at a later stage, prior to the divergence of mosses. The GR1 homolog from Physcomitrella carries a PTS1-related tripeptide, LKV>, of weak predicted targeting strength, similar to TNL> in higher plants. Notably, leucine (L) has recently been experimentally verified as another allowed residue at pos. -3 in plant PTS1 tripeptides (Lingner T, Antonicelli GE, Kataya A, Reumann S, unpubl. data).
The most intriguing question of multiple protein targeting concerns the possible existence and identity of the underlying regulatory mechanism. Two mechanisms that regulate protein distribution between the cytosol and peroxisomes can be envisioned: (1) a rather static mechanism of protein distribution that is largely determined by the PTS strength, i.e., the affinity of the PTS1 domain to its receptor, PEX5p, and (2) a dynamic mechanism of post-transcriptional or post-translational nature that allows adequate responses to changing demands in GR1 compartmentalization. Notably, even in the case of a rather simple regulation of protein distribution by PTS1 strength, the peroxisome import rate by weak PTS1s in vivo depends on the rate of gene expression; low-abundance proteins with weak PTS1s may still be imported quantitatively into peroxisomes, but not high-abundance proteins. Therefore, the low peroxisome import rate of GR1 upon expression from the strong 35S promoter does not allow the conclusion to be made that GR1 is largely cytosolic in vivo. This question needs to be addressed by expressing N-terminally or internally tagged GR1 versions, preferentially in a gr1 knockout background, from the native GR1 promoter. The fact that GR1 was identified in leaf peroxisomes that were isolated from plants grown under standard conditionsCitation13,Citation14 supports the idea of a constitutive role of GR1 in peroxisomes.
In light of the strong need and major function of the peroxisomal ascorbateglutathione cycle under sudden conditions of catalase inactivation,Citation25 it is reasonable to postulate that the ratio of GR1 distribution between peroxisomes and the cytosol is dynamic and adjustable depending on H2O2 overproduction in the matrix by post-transcriptional and/or post-translational mechanisms. Interestingly, pea (and later, Arabidopsis) GR2 was one of the first proteins shown to be dually targeted to two plant cell compartments, mitochondria and plastids.Citation12,Citation26 Dual targeting of GR1/2 thus emerges as a specific property of the enzyme. The regulatory mechanism(s) underlying dual targeting of both GR1 and GR2 are still unknown and remain to be studied. The exclusively cytosolic localization of EYFP-GR1 in tobacco protoplasts () may indicate that peroxisome targeting is prevented in this expression system by PTS1 inaccessibility caused by conformational changes or binding of an accessory protein that regulates enzyme distribution between the cytosol and peroxisomes.
Conclusions
The data presented here conclusively identify the peroxisomal GR isoform in the first plant species at the molecular level, thereby completing the ascorbate-glutathione cycle in plant peroxisomes. In vivo studies, for instance, reverse genetics, as have been recently initiated for cytosolic gr1,Citation27 need to follow in order to investigate the functions of cytosolic and peroxisomal GR1 in planta and “in organello.” Future studies will also need to focus on the regulatory mechanism of dual targeting.
Abbreviations
| aa | = | amino acid |
| APX | = | ascorbate peroxidase |
| CaMV | = | cauliflower mosaic virus |
| CFP | = | cyan fluorescent protein |
| DHAR | = | dehydroascorbate reductase |
| EST | = | expressed sequence tag |
| GR | = | glutathione reductase |
| MDAR | = | monodehydroascorbate reductase |
| PTD | = | peroxisome targeting domain |
| PTS1/2 | = | peroxisome targeting signal type 1/2 |
| YFP | = | yellow fluorescent protein |
Figures and Tables
Figure 1 Analysis of PTS1 conservation in plant GR1 homologs. Sequences of full-length protein (FLP) plant GR1 homologs or ESTs (“EST”) were identified by BLAST and phylogenetic analysis, aligned by ClustalX, and conserved residues were shaded by Genedoc. In addition to spermatophyta, homologs from bryophyta and chlorophyta were analyzed for PTS1 conservation. For a phylogenetic analysis of the full-length proteins, see also Supplementary . The species abbreviations are as follows: Aa, Artemisia annua; At, Arabidopsis thaliana; Bn, Brassica napus; Br, Brassica rapa; Ci, Cichorium intybus; Cr, Chlamydomonas reinhardtii; Cs, Cynara scolymus; Fv, Fragaria vesca; Ha, Helianthus annuus; Msp, Micromonas sp. RCC 299; Mt, Medicago truncatula; Nt, Nicotiana tabacum; Os, Oryza sativa; Pk, Picrorhiza kurrooa; Ppat, Physcomitrella patens subsp. patens; Ps, Pisum sativum; Ptri, Populus trichocarpa; Rc, Ricinus communis; Rs, Raphanus sativus; Tp, Trifolium pratense; Tpus, Triphysaria pusilla; Vc, Volvox carteri f. nagariensis; Vv, Vitis vinifera; Zm, Zea mays.
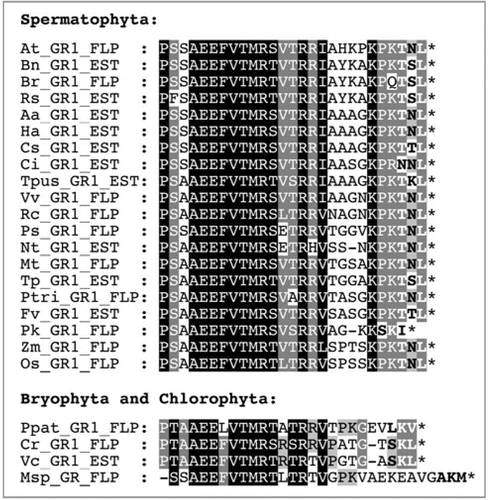
Figure 2 In vivo subcellular targeting analysis of Arabidopsis GR1. The full-length cDNA of Arabidopsis GR1 was fused N-terminally with the reporter protein EYFP and expressed transiently in onion epidermal cells upon biolistic bombardment or in tobacco protoplasts upon polyethylene glycol-mediated transformation. To characterize TN L> as the PTS1 of GR1, EYFP was extended C-terminally by the predicted peroxisome targeting domain of GR1, comprising the C-terminal 10 aa residues (AHKPKPKTNL>). In double transformants, peroxisomes were labeled with gMD H-CFP,Citation28 and the cyan fluorescence was converted to red to facilitate the detection of green and cyan fluorescent peroxisomes as yellow organelles in image overlays (merge). EYFP alone and EYFP extended C-terminally by 10 glycine residues (EYFP-10G) served as negative controls to verify the absence of endogenous bacteria. Scale bar: 10 µm.
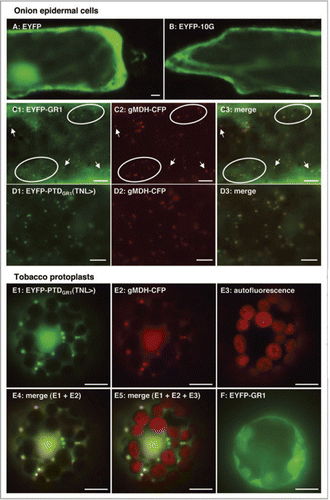
Figure 3 Model of the evolutionary development of the peroxisomal and cytosolic functions of GR1 in spermatophyta. The canonical nature and high peroxisome targeting strength of PTS1s in GR homologs of chlorophyta (Chlamydomonas, Volvox and Micromonas) indicate an exclusive compartmentalization of GR1 in peroxisomes in green algae. By contrast, the non-canonical nature and predicted (LKV>) and proven (TN L>) low peroxisome targeting strength in Physcomitrella and higher plants, respectively, indicate dual enzyme targeting to peroxisomes and the cytosol in bryophyta and spermatophyta.
Additional material
Download Zip (88 KB)Acknowledgements
We would like to thank the Arabidopsis Biological Resource Center (ABRC) for providing the GR1 full-length cDNA and Aline Benichou and Dr. Xiong-Yan Chen for experimental support.
Addendum to:
References
- Huang AHC, Moore TS, Trelease RN. Plant peroxisomes 1983; New York Academic Press
- Halliwell B, Foyer CH. Ascorbic acid, metal ions and the superoxide radical. Biochem J 1976; 155:697 - 700
- del Rio LA, Sandalio LM, Corpas FJ, Palma JM, Barroso JB. Reactive oxygen species and reactive nitrogen species in peroxisomes. Production, scavenging and role in cell signaling. Plant Physiol 2006; 141:330 - 335
- Noctor G, Foyer CH. ASCORBATE AND GLUTATHIONE: Keeping Active Oxygen Under Control. Annu Rev Plant Physiol Plant Mol Biol 1998; 49:249 - 279
- Romero-Puertas MC, Corpas FJ, Sandalio LM, Leterrier M, Rodriguez-Serrano M, Del Rio LA, et al. Glutathione reductase from pea leaves: response to abiotic stress and characterization of the peroxisomal isozyme. New Phytol 2006; 170:43 - 52
- Jimenez A, Hernandez JA, Del Rio LA, Sevilla F. Evidence for the presence of the ascorbate-glutathione cycle in mitochondria and peroxisomes of pea leaves. Plant Physiol 1997; 114:275 - 284
- Jimenez A, Hernandez JA, Pastori G, del Rio LA, Sevilla F. Role of the ascorbate-glutathione cycle of mitochondria and peroxisomes in the senescence of pea leaves. Plant Physiol 1998; 118:1327 - 1335
- Palma JM, Jimenez A, Sandalio LM, Corpas FJ, Lundqvist M, Gomez M, et al. Antioxidative enzymes from chloroplasts, mitochondria and peroxisomes during leaf senescence of nodulated pea plants. J Exp Bot 2006; 57:1747 - 1758
- Zhang H, Wang J, Nickel U, Allen RD, Goodman HM. Cloning and expression of an Arabidopsis gene encoding a putative peroxisomal ascorbate peroxidase. Plant Mol Biol 1997; 34:967 - 971
- Leterrier M, Corpas FJ, Barroso JB, Sandalio LM, del Rio LA. Peroxisomal monodehydroascorbate reductase. Genomic clone characterization and functional analysis under environmental stress conditions. Plant Physiol 2005; 138:2111 - 2123
- Lisenbee CS, Lingard MJ, Trelease RN. Arabidopsis peroxisomes possess functionally redundant membrane and matrix isoforms of monodehydroascorbate reductase. Plant J 2005; 43:900 - 914
- Chew O, Whelan J, Millar AH. Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J Biol Chem 2003; 278:46869 - 46877
- Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, Antonicelli GE, et al. Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways and defense mechanisms. Plant Cell 2007; 19:3170 - 3193
- Reumann S, Quan S, Aung K, Yang P, Manandhar-Shrestha K, Holbrook D, et al. In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol 2009; 150:125 - 143
- Stevens RG, Creissen GP, Mullineaux PM. Cloning and characterisation of a cytosolic glutathione reductase cDNA from pea (Pisum sativum L.) and its expression in response to stress. Plant Mol Biol 1997; 35:641 - 654
- Reumann S. Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol 2004; 135:783 - 800
- Nowak K, Luniak N, Witt C, Wustefeld Y, Wachter A, Mendel RR, et al. Peroxisomal localization of sulfite oxidase separates it from chloroplast-based sulfur assimilation. Plant Cell Physiol 2004; 45:1889 - 1894
- Mullen RT, Lee MS, Flynn CR, Trelease RN. Diverse amino acid residues function within the type 1 peroxisomal targeting signal. Implications for the role of accessory residues upstream of the type 1 peroxisomal targeting signal. Plant Physiol 1997; 115:881 - 889
- Dixon DP, Hawkins T, Hussey PJ, Edwards R. Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily. J Exp Bot 2009; 60:1207 - 1218
- Noctor G, Gomez L, Vanacker H, Foyer CH. Interactions between biosynthesis, compartmentation and transport in the control of glutathione homeostasis and signalling. J Exp Bot 2002; 53:1283 - 1304
- Carrie C, Murcha MW, Kuehn K, Duncan O, Barthet M, Smith PM, et al. Type II NAD(P)H dehydrogenases are targeted to mitochondria and chloroplasts or peroxisomes in Arabidopsis thaliana. FEBS Lett 2008; 582:3073 - 3079
- Carrie C, Kuhn K, Murcha MW, Duncan O, Small ID, O’Toole N, et al. Approaches to defining dualtargeted proteins in Arabidopsis. Plant J 2009; 57:1128 - 1139
- Wimmer B, Lottspeich F, van der Klei I, Veenhuis M, Gietl C. The glyoxysomal and plastid molecular chaperones (70-kDa heat shock protein) of watermelon cotyledons are encoded by a single gene. Proc Natl Acad Sci USA 1997; 94:13624 - 13629
- Gebhardt JS, Wadsworth GJ, Matthews BF. Characterization of a single soybean cDNA encoding cytosolic and glyoxysomal isozymes of aspartate aminotransferase. Plant Mol Biol 1998; 37:99 - 108
- Feierabend J, Engel S. Photoinactivation of catalase in vitro and in leaves. Arch Biochem Biophys 1986; 251:567 - 576
- Creissen G, Reynolds H, Xue Y, Mullineaux P. Simultaneous targeting of pea glutathione reductase and of a bacterial fusion protein to chloroplasts and mitochondria in transgenic tobacco. Plant J 1995; 8:167 - 175
- Marty L, Siala W, Schwarzlander M, Fricker MD, Wirtz M, Sweetlove LJ, et al. The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. Proc Natl Acad Sci USA 2009; 106:9109 - 9114
- Ma C, Haslbeck M, Babujee L, Jahn O, Reumann S. Identification and characterization of a stressinducible and a constitutive small heat-shock protein targeted to the matrix of plant peroxisomes. Plant Physiol 2006; 141:47 - 60