Abstract
Several proteomic studies in Arabidopsis have shown the presence of heterogeneous ribosomal populations in different tissues. However, the phenotypic consequences of the imbalance of those ribosomal populations, and the regulatory mechanisms activated to control specific ratios between them, have yet to be evaluated. In our previous report, the phenotypic characterization of the Arabidopsis ribosomal L4 (RPL4) family suggests that the maintenance of proper auxin-regulated developmental responses requires the simultaneous presence of RPL4A- and RPL4D-containing ribosomes. Based on the analysis of the compensatory mechanisms within the RPL4 family proteins in the rpl4a and rpl4d backgrounds, we propose the Gene Dosage Balance Hypothesis (GDBH) as a regulatory mechanism for ribosomal complexes in Arabidopsis. By using the concepts of dosage compensation and hierarchy, GDBH is able to explain the severity and specificity of different ribosomal mutant phenotypes associated with the same ribosomal complex.
The Arabidopsis ribosome is a complex structure whose assembly requires the proper temporal and spatial organization of four RNA species and 81 ribosomal proteins encoded by large multigene families.Citation1,Citation2 Although the presence of multigene families encoding ribosomal proteins suggests that the system maintains redundancy, several studies have shown that genetic defects in individual ribosomal components can cause deleterious developmental effects in a gene dosage dependent manner.Citation3–Citation6 Since the loss of a single Arabidopsis ribosomal protein affects the performance of the whole ribosomal complex, and this effect has been observed in different ribosomal protein mutants (RPMs),Citation6 we questioned whether there is a common regulatory mechanism involving the different components of the ribosomal complex in Arabidopsis.
In yeast, the gene dosage effects of different elements belonging to protein complexes have been explained by a mechanistic model named the Gene Dosage Balance Hypothesis (GDBH).Citation7 According to the GDBH, the stoichiometric rules that govern the assembly of proteins in a complex are essential for the function of the whole system and any deviation from the normal stoichiometry would be deleterious; thus, both underexpression and overexpression of individual components can potentially lower the fitness of the organism.Citation7–Citation11 The GDBH has been applied to different organisms and examples of complexes where underexpression of a single component is harmful include mutations in Drosophila melanogaster ribosomal genes that cause the Minute phenotype,Citation12 and the quantitative reduction in synthesis of the human ribosomal protein S4 observed in individuals with Turner syndrome.Citation13 Alternatively, examples of complexes where the overexpression of a single component has deleterious effects include the yeast genes GPA1, STE4 and STE18 which encode homologs of the mammalian G protein α, β and γ subunits respectively,Citation14 and the Snf1/AMP-activated protein kinase complex involved in stress response.Citation15 In this addendum we will discuss the application of the GDBH to the Arabidopsis ribosomal complexes and its consequences in the auxin-related phenotypes observed in different RPMs.
In Arabidopsis, different RPMs share impaired auxin-dependent responses but their phenotypic consequences differ in terms of severity.Citation6 Instead of being considered quantitative traits, those phenotypic differences has been described as “specific” and widely used to sort and characterize each Arabidopsis RPM. Accordingly, the RPM stv1 has been characterized by its aberrant gynaeceum development,Citation16 the RPMs rpl10a and rpl9 by their role in leaf patterning control,Citation17 the RPMs rpl28a and rpl5a, by their role in leaf polarity,Citation18 and the RPMs rpl4 by their vacuolar trafficking responses.Citation6
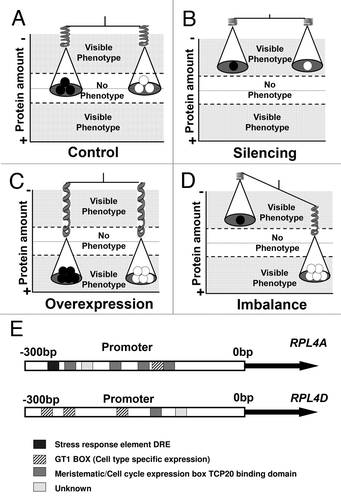
From the regulatory point of view, the results from Rosado et al.Citation6 support the GDBH as a plausible explanation for auxin-related phenotypes observed in different RPMs. In that study, the rpl4 mutations activate compensatory mechanisms that include the transcriptional and translational activation of the alternative ribosomal RPL4 family member. As a consequence, the amount of total RPL4 protein in different rpl4 mutant backgrounds is higher than in wild-type; however, the ratio of RPL4A:RPL4D in those backgrounds is imbalanced. Since the phenotypes found in our rpl4 imbalanced lines resemble several recessive RPMs,Citation3,Citation5,Citation16–Citation18 ribosomal silenced lines,Citation19 and semi-dominant ribosomal mutations,Citation20 we hypothesize that the phenotype specificity and severity of the different RPMs might be a consequence of a regulatory imbalance of the entire ribosomal complex as proposed by the GDBH (). In this model, the outcome of deletions or duplications of genes encoding ribosomal components depends on their respective topological positions and mechanism of assembly into the complex.Citation7,Citation10 Thus, the independent spatial and temporal regulatory characteristics of each member of the r-protein families in Arabidopsis () as well as its hierarchical position within the ribosome might trigger specific compensatory mechanisms to cope with the absence of particular ribosomal proteins. Consequently, the severity and/or specificity of the auxin-related phenotypes in different RPMs described in the literature could be explained as the relative success of the remaining ribosomal proteins to coordinate their responses to assemble optimally functional ribosomes.
In conclusion, we postulate that GBDH is a regulatory mechanism that explains the severity and specificity of the auxin-related phenotypes observed in many Arabidopsis RPMs.
Figures and Tables
Figure 1 Schematic representation of the Gene Dosage Balance Hypothesis applied to a hypothetic two member ribosomal protein family in Arabidopsis. (A) A wild type plant maintains the correct amount of protein and the balance among different family members. As a consequence, the equilibrium position is located in the middle of the graph (white) and no phenotypes are observed. (B) A silenced line maintains the balance among different ribosomal protein family members but fails to obtain optimal protein amounts. As a consequence, the equilibrium position is displaced to the upper area of the graph (grey) and aberrant phenotypes are observed. (C) A ribosomal overexpression line might maintain the balance among different family members, but the excess of ribosomal proteins displace the equilibrium position to the lower area of the graph (grey). As a consequence, aberrant phenotypes are observed. (D) An imbalanced line might have the sufficient protein amounts but fails to maintain the proportions among the different protein family members. As a consequence, the equilibrium position is displaced to both, the upper and lower areas of the graph (grey) and aberrant phenotypes are observed. The two arms of the scale represent different equilibrium positions, and the numbers of white a black circles represent the relative ribosomal protein amounts of two different ribosomal family members. Relative protein amounts are indicated as a continuum with increased (+) and decreased (−) shown on the Y-axis in this model. (E) The different regulatory boxes present in the promoter region of the RPL4 family members might act as a fine tuning mechanism to trigger specific compensatory mechanisms as a response to different ribosomal protein mutations. The regulatory boxes are indicated with different color patterns and were identified using ATHENA.Citation21
Acknowledgements
We thank Dr. Glenn Hicks (University of California, Riverside) for his critical reading of the manuscript. This work was funded by Department of Energy, Division of Energy Biosciences, Grant DE-FG03-02ER15295/A000.
Addendum to:
References
- Barakat A, Szick-Miranda K, Chang IF, Guyot R, Blanc G, Cooke R, et al. The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol 2001; 127:398 - 415
- Giavalisco P, Wilson D, Kreitler T, Lehrach H, Klose J, Gobom J, et al. High heterogeneity within the ribosomal proteins of the Arabidopsis thaliana 80S ribosome. Plant Mol Biol 2005; 57:577 - 591
- Ito T, Kim GT, Shinozaki K. Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J 2000; 22:257 - 264
- Tsugeki R, Kochieva EZ, Fedoroff NV. A transposon insertion in the Arabidopsis SSR16 gene causes an embryo-defective lethal mutation. Plant J 1996; 10:479 - 489
- Van Lijsebettens M, Vanderhaeghen R, De Block M, Bauw G, Villarroel R, Van Montagu M. An S18 ribosomal protein gene copy at the Arabidopsis PFL locus affects plant development by its specific expression in meristems. EMBO J 1994; 13:3378 - 3388
- Rosado A, Sohn EJ, Drakakaki G, Pan S, Swidergal A, Xiong Y, et al. Auxin-mediated ribosomal biogenesis regulates vacuolar trafficking in Arabidopsis. Plant Cell 2010; 22:143 - 158
- Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature 2003; 424:194 - 197
- Veitia RA. Nonlinear effects in macromolecular assembly and dosage sensitivity. J Theor Biol 2003; 220:19 - 25
- Veitia RA. Paralogs in polyploids: one for all and all for one?. Plant Cell 2005; 17:4 - 11
- Veitia RA, Bottani S, Birchler JA. Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet 2008; 24:390 - 397
- Birchler JA, Bhadra U, Bhadra MP, Auger DL. Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes and quantitative traits. Dev Biol 2001; 234:275 - 288
- Lambertsson A. The minute genes in Drosophila and their molecular functions. Adv Genet 1998; 38:69 - 134
- Zinn AR, Ross JL. Turner syndrome and haploinsufficiency. Current Opinion in Genetics & Development 1998; 8:322 - 327
- Dohlman HG, Thorner JW. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu Rev Biochem 2001; 70:703 - 754
- Vincent O, Townley R, Kuchin S, Carlson M. Subcellular localization of the Snf1 kinase is regulated by specific beta subunits and a novel glucose signaling mechanism. Genes Dev 2001; 15:1104 - 1114
- Nishimura T, Wada T, Yamamoto KT, Okada K. The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell 2005; 17:2940 - 2953
- Pinon V, Etchells JP, Rossignol P, Collier SA, Arroyo JM, Martienssen RA, et al. Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development 2008; 135:1315 - 1324
- Yao Y, Ling Q, Wang H, Huang H. Ribosomal proteins promote leaf adaxial identity. Development 2008; 135:1325 - 1334
- Degenhardt RF, Bonham-Smith PC. Arabidopsis ribosomal proteins RPL23aA and RPL23aB are differentially targeted to the nucleolus and are desperately required for normal development. Plant Physiol 2008; 147:128 - 142
- Weijers D, Franke-van Dijk M, Vencken RJ, Quint A, Hooykaas P, Offringa R. An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 2001; 128:4289 - 4299
- O'Connor TR, Dyreson C, Wyrick JJ. Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 2005; 21:4411 - 4413