Abstract
Alteration of gene expression plays a central role in the transmission of developmental and environmental signals. The steady-state transcript level within a cell is determined by the combination of the rate synthesis and the rate of degradation. While altering the rate of mRNA turnover is known to provide a rapid mechanism to reprogram transcript levels, research has largely focused on changes in transcriptional regulation as a mechanism to control mRNA levels. However, recent studies have begun to explore the role of mRNA decay in reprogramming the transcriptome.
Introduction
In eukaryotic organisms, the rate limiting step in mRNA degradation is shortening of the poly(A) tail, a process referred to as deadenylation.Citation1–Citation3 Once a transcript has been deadenylated, degradation proceeds via two distinct pathways. In one pathway transcripts are degraded in a 3′-to-5′ manner via the exosome.Citation2,Citation4 Alternatively, deadenylated mRNA can also undergo 5′-to-3′ decay. The 5′-to-3′ pathway first requires that the 5′ guanine nucleotide cap be removed by a decapping complex consisting of DCP1 and DCP2. The decapped mRNA are then degraded by the exonuclease XRN1.Citation2,Citation5
Numerous studies, largely conducted in yeast, have yielded considerable mechanistic insight into the process and players involved in deadenylation. Two of the best characterized deadenylases, CAF1 and CCR4, have been found to be present and active in a wide range of organisms.Citation6–Citation12 In yeast, the CCR4-CAF1 complex is composed of nine subunits: CCR4, CAF1, NOT1-5, CAF130 and CAF40.Citation11 Humans have multiple CCR4 and CAF1 homologs, creating at least four complex variants that consist of seven stable core proteins and mutually exclusive deadenylase subunits.Citation13 In addition to the CCR4-CAF1 complex both poly(A) ribonuclease (PARN), for which mutants in Arabidopsis are embryo-lethal, and poly(A) nuclease (PAN) are also active deadenylases.Citation1,Citation2,Citation14,Citation15
Phenotypic analysis of caf1 mutants has been conducted in a range of model organisms, demonstrating that CAF1 is vital for a wide range of biological processes. In yeast, caf1 deletion strains are sensitive to high temperatures and caffeine.Citation16 siRNA mediated knock-down of two CAF1 paralogs (CNOT7 and CNOT8) in human breast cancer cells results in a reduced cell proliferation rate and an increase in G1-phase cells.Citation17 Analysis of CAF1 in C. elegans using RNAi and deletion alleles demonstrates that CAF1 is essential for embryonic and larval development.Citation18 Additionally, CAF1 is required for normal growth of typanosomes.Citation9 Loss of CAF1 in mice leads to sterility, demonstrating that CAF1 also plays a role in reproductive development.Citation19 One study found that overexpression of a pepper CAF1 homolog (Capsicum annuum; CaCAF1) in tomato resulted in growth enhancement and resistance to the oomycete pathogen Phytophthora infestans. Conversely, when CaCAF1 is silenced in pepper there is significant growth retardation and plants are susceptible to the pathogen Xanthomonas axonopodis pv. Vesicatoria.Citation20 Finally, recent work investigating the function of CAF1 proteins in Arabidopsis has been conducted and will be the focus of the remainder of this review.
CAF1 Family in Plants
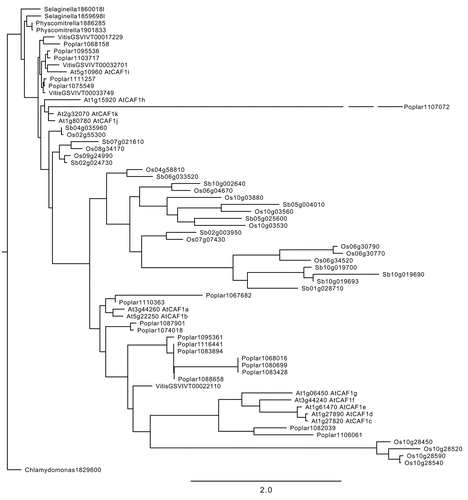
A wide survey of CAF1 orthologs among eukaryotes demonstrates that most species (including yeast, trypanosomes, worms, flies, mice and humans) contain only one or two CAF1 genes.Citation9,Citation10,Citation18,Citation19,Citation21–Citation23 In contrast, the CAF1 gene family has significantly expanded in angiosperms. We uncovered CAF1 homologs using a Basic Local Alignment Search Tool (BLAST) of fully sequenced plant genomes at phytozome (www.phytozome.net) ( and Walley et al.Citation24). Specifically, we found one closely related sequence in algae (Chlamydomonas reinhardtii), two in moss and lycophytes (Physcomitrella patens and Selaginella moellendorffii), four in grape (Vitis vinifera), twelve in Sorghum (Sorghum bicolor), sixteen sequences in rice (Oryza sativa), nineteen sequences in poplar (Populus trichocarpa), and eleven in Arabidopsis. Whether all of these homologs encode active deadenylases remains to be determined. However, the RNaseD domain responsible for exonuclease activityCitation22 is well conserved. Furthermore, the two Arabidopsis homologs examined, AtCAF1a and AtCAF1b, exhibit deadenylase activity in vitro and partially complement growth defects of a yeast caf1 deletion strain.Citation7 The expansion in the angiosperm CAF1 family suggests that sub- or neo-functionalization may have occurred enabling individual CAF1 proteins to function in specific instances, such as in response to stress and/or developmental cues.
AtCAF1s Mediate Stress Responses
Two AtCAF1 genes were first implicated in stress signaling via transcript profiling experiments. These experiments identified AtCAF1a and AtCAF1b as responding transiently to a number of hormones and stresses including jasmonic acid (JA), wounding, cold and Botrytis cinerea.Citation25–Citation28 Furthermore, Ma and BohnertCitation29 classified AtCAF1b as a universal stress response gene following analysis of the AtGenExpress stress response data. Additionally, AtCAF1a and AtCAF1b, assayed by northern blot, are induced rapidly and transiently in response to MeJA, ABA, ACC, SA, wounding as well as Pseudomonas syringae.Citation7 Targeted analysis of AtCAF1 genes in response to wounding using RT-qPCR revealed that a subset exhibit differential expression patterns in response to mechanical wounding.Citation24 Consistent with their induction by a range of stresses, Atcaf1a and Atcaf1b mutants have reduced tolerance to virulent Pseudomonas syringae pv tomato DC3000 as well as the reactive oxygen species (ROS) inducer methyl viologen (MV; paraquat).Citation7,Citation24 In contrast, neither Atcaf1a nor Atcaf1b exhibited altered tolerance to water-logging or the necrotrophic pathogen Botrytis cinerea.Citation24 Intriguingly, Atcaf1a, but not Atcaf1b, mutants exhibit increased tolerance to 200 mM NaCl.Citation24 The observed difference in salt tolerance represents functional specificity between these two paralogs, which is consistent with a sub (or neo) functionalization event, as suggested by the expansion of the CAF1 family in angiosperms.
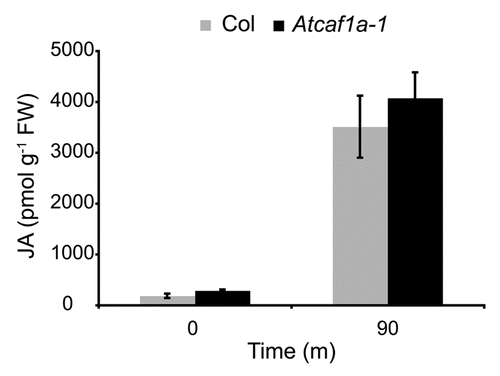
The induction of AtCAF1a and AtCAF1b by both JA and wounding suggests their involvement in JA signaling. In a recent study, Liang et al.Citation7 report that AtCAF1a and AtCAF1b deadenylate transcripts of JA biosynthetic and JA responsive genes. Specifically, they report that Atcaf1a and Atcaf1b mutants are defective in deadenylation of LOX2, VSP1 and CHIB following induction by wounding, JA or ACC treatment, respectively. These results support the notion that AtCAF1a and AtCAF1b are indeed required for proper JA signaling. However, we monitored JA levels in Atcaf1a-1 mutants and did not detect any difference from WT at 0 or 90 min post wounding (). It should be noted that the timepoints we assayed were earlier than Liang et al.Citation7 who report differences in LOX2 deadenylation 3–15 h post wounding. It would therefore be of interest to determine if JA levels are perturbed in Atcaf1a at later timepoints.
AtCAF1a and AtCAF1b can Target Specific Transcripts
The increased tolerance of Atcaf1a, but not Atcaf1b, to NaCl suggests that these two deadenylases are capable of targeting specific mRNAs. Multiple lines of evidence at the molecular level support this hypothesis.Citation24 Transcript profiling of Atcaf1a and Atcaf1b results in largely distinct sets of altered transcripts between the two mutants. Furthermore, examination of the mis-expressed transcripts in response to wounding reveals limited overlap between timepoints assayed (ex. Atcaf1a at 0 min vs. Atcaf1a at 30 min), suggesting that these deadenylases act with temporal specificity as well. Perhaps the strongest evidence for substrate specificity comes from analysis of AtPI4Kγ3, which is specifically increased in abundance in Atcaf1b. Analysis of the deadenylation state of AtPI4Kγ3 mRNA, via poly(A) tail length (PAT) assays,Citation30 demonstrates that the poly(A) tail length increases specifically in the absence of AtCAF1b.
While the above experiments establish that AtCAF1a and AtCAF1b are capable of targeting specific mRNAs the question of how this specificity is achieved remains to be addressed. Evidence from experiments conducted in other eukaryotes suggests that targeting of AtCAF1a and AtCAF1b to particular mRNA(s) may be facilitated via interaction between specific AtCAF1 proteins and RNA binding proteins. In Drosophila, physical interaction between CAF1 and the RNA binding protein Smaug enables the CCR4-CAF1 complex to specifically target Hsp83 and nanos mRNAs.Citation31,Citation32 A similar mechanism exists in yeast, where the RNA binding protein Mpt5p directly interacts with CAF1 thereby recruiting the CCR4-CAF1 complex specifically to HO mRNA.Citation33 Given the high degree of conservation among eukaryotic CAF1 homologs it is likely that this targeting mechanism may be similarly utilized in plants.
Conclusions and Perspectives
In conclusion, phylogenic analysis suggests that the CAF1 family has expanded in angiosperms. This expansion may provide a robust combinatorial framework for individual AtCAF1 proteins to target specific mRNA substrates in response to developmental and environmental signals. The ability of AtCAF1s to target specific transcripts is exemplified by AtCAF1a and AtCAF1b at both the molecular and organismal level. Biochemical characterization of AtCAF1a and AtCAF1b associated mRNA binding proteins should help elucidate how they capable of targeting specific mRNA substrates.
Abbreviations
| CCR4 | = | carbon catabolite repressor 4 |
| CAF1 | = | CCR4 associated factor1 |
| PARN | = | poly(A) ribonuclease |
| PAN | = | poly(A) nuclease |
| JA | = | jasmonic acid |
| MeJA | = | methyl jasmonate |
| ABA | = | abscisic acid |
| ACC | = | 1-aminocyclopropane-1-carboxylic acid |
| SA | = | salicylic acid |
| RT-qPCR | = | quantitative reverse-transcriptase-polymerase chain reaction |
| AtPI4Kγ | = | putative phosphatidylinositol 4-kinase |
| ROS | = | reactive oxygen species |
| MV | = | methyl viologen |
Figures and Tables
Acknowledgements
This work was supported by NSF grants 0543904 and 0606838 awarded to K.D.
References
- Meyer S, Temme C, Wahle E. Messenger RNA Turnover in Eukaryotes: Pathways and Enzymes. Crit Rev Biochem Mol Biol 2004; 39:197 - 216
- Belostotsky DA, Sieburth LE. Kill the messenger: mRNA decay and plant development. Curr Opin Plant Biol 2009; 12:96 - 102
- Chiba Y, Green PJ. mRNA Degradation Machinery in Plants. J Plant Biol 2009; 52:114 - 124
- Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, et al. Genome-Wide High-Resolution Mapping of Exosome Substrates Reveals Hidden Features in the Arabidopsis Transcriptome. Cell 2007; 131:1340 - 1353
- Jiao Y, Riechmann JL, Meyerowitz EM. Transcriptome-Wide Analysis of Uncapped mRNAs in Arabidopsis Reveals Regulation of mRNA Degradation. Plant Cell 2008; tpc.108.062786
- Morozov IY, Jones MG, Razak AA, Rigden DJ, Caddick MX. CUCU Modification of mRNA Promotes Decapping and Transcript Degradation in Aspergillus nidulans. Mol Cell Biol 2010; 30:460 - 469
- Liang W, Li C, Liu F, Jiang H, Li S, Sun J, et al. The Arabidopsis homologs of CCR4-associated factor 1 show mRNA deadenylation activity and play a role in plant defence responses. Cell Res 2009; 19:307 - 316
- Mauxion F, Faux C, Seraphin B. The BTG2 protein is a general activator of mRNA deadenylation. EMBO J 2008; 27:1039 - 1048
- Schwede A, Ellis L, Luther J, Carrington M, Stoecklin G, Clayton C. A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucl Acids Res 2008; gkn108
- Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J 2004; 23:2862 - 2871
- Cui Y, Ramnarain D, Chiang Y-C, Ding L-H, McMahon J, Denis C. Genome wide expression analysis of the CCR4-NOT complex indicates that it consists of three modules with the NOT module controlling SAGA-responsive genes. Mol Genet Genomics 2008; 279:323 - 337
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The Transcription Factor Associated Ccr4 and Caf1 Proteins Are Components of the Major Cytoplasmic mRNA Deadenylase in Saccharomyces cerevisiae. Cell 2001; 104:377 - 386
- Lau NC, Kolkman A, van Schaik FMA, Mulder KW, Pijnappel WWMP, Heck AJR, et al. Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem J 2009; 422:443 - 453
- Reverdatto SV, Dutko JA, Chekanova JA, Hamilton DA, Belostotsky DA. mRNA deadenylation by PARN is essential for embryogenesis in higher plants. RNA 2004; 10:1200 - 1214
- Chiba Y, Johnson MA, Lidder P, Vogel JT, van Erp H, Green PJ. AtPARN is an essential poly(A) ribonuclease in Arabidopsis. Gene 2004; 328:95 - 102
- Hata H, Mitsui H, Liu H, Bai Y, Denis CL, Shimizu Y, et al. Dhh1p, a Putative RNA Helicase, Associates with the General Transcription Factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics 1998; 148:571 - 580
- Aslam A, Mittal S, Koch F, Andrau J-C, Winkler GS. The Ccr4-Not Deadenylase Subunits CNOT7 and CNOT8 Have Overlapping Roles and Modulate Cell Proliferation. Mol Biol Cell 2009; 20:3840 - 3850
- Molin L, Puisieux A. C. elegans homologue of the Caf1 gene, which encodes a subunit of the CCR4-NOT complex, is essential for embryonic and larval development and for meiotic progression. Gene 2005; 358:73 - 81
- Nakamura T, Yao R, Ogawa T, Suzuki T, Ito C, Tsunekawa N, et al. Oligo-astheno-teratozoospermia in mice lacking Cnot7, a regulator of retinoid X receptor beta. Nat Genet 2004; 36:528 - 533
- Sujon S, Hyun Woo O, Hye Sun C, Kwang-Hyun B, Eun Soo S, Young Hee J, et al. Capsicum annuum CCR4-associated factor CaCAF1 is necessary for plant development and defence response. Plant J 2007; 51:792 - 802
- Yamashita A, Chang T-C, Yamashita Y, Zhu W, Zhong Z, Chen C-YA, et al. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 2005; 12:1054 - 1063
- Daugeron M-C, Mauxion F, Seraphin B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucl Acids Res 2001; 29:2448 - 2455
- Albert TK, Lemaire M, van Berkum NL, Gentz R, Collart MA, Timmers HTM. Isolation and characterization of human orthologs of yeast CCR4-NOT complex subunits. Nucl Acids Res 2000; 28:809 - 817
- Walley JW, Kelley DR, Nestorova G, Hirschberg DL, Dehesh K. Arabidopsis deadenylases AtCAF1a and AtCAF1b play overlapping and distinct roles in mediating environmental stress responses. Plant Physiol 2009; pp.109.149005
- Zheng W, Zhai Q, Sun J, Li C-B, Zhang L, Li H, et al. Bestatin, an Inhibitor of Aminopeptidases, Provides a Chemical Genetics Approach to Dissect Jasmonate Signaling in Arabidopsis. Plant Physiol 2006; 141:1400 - 1413
- Ferrari S, Galletti R, Denoux C, De Lorenzo G, Ausubel FM, Dewdney J. Resistance to Botrytis cinerea Induced in Arabidopsis by Elicitors Is Independent of Salicylic Acid, Ethylene, or Jasmonate Signaling But Requires PHYTOALEXIN DEFICIENT3. Plant Physiol 2007; 144:367 - 379
- Walley JW, Coughlan S, Hudson ME, Covington MF, Kaspi R, Banu G, et al. Mechanical Stress Induces Biotic and Abiotic Stress Responses via a Novel cis-Element. PLoS Genet 2007; 3:172
- Lee B-h, Henderson DA, Zhu J-K. The Arabidopsis Cold-Responsive Transcriptome and Its Regulation by ICE1. Plant Cell 2005; 17:3155 - 175
- Ma S, Bohnert H. Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures and cell-specific expression. Genome Biol 2007; 8:49
- Salles FJ, Strickland S. Haynes S. Analysis of poly(A) tail lengths by PCR: The PAT assay. Methods in Molecular Biology 1999; Totowa N.J. Humana Press 441 - 448
- Zaessinger S, Busseau I, Simonelig M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development 2006; 133:4573 - 4583
- Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA. Smaug Recruits the CCR4/POP2/NOT Deadenylase Complex to Trigger Maternal Transcript Localization in the Early Drosophila Embryo. Curr Biol 2005; 15:284 - 294
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol 2006; 13:533 - 539