Abstract
Genes coding for drought response element binding (DREB) proteins regulate transcription of a large number of downstream genes involved in the plant response to abiotic stresses. However the regulation of DREB genes themselves is not well understood. Using a bioinformatics approach, we identified the over-represented motifs in promoters of DREB genes of sorghum and rice as compared to all the other promoters in their genomes. Aligned orthologous promoter pairs of sorghum and rice DREBs were then used to identify co-localized motifs from among the over-represented ones, assuming that such motifs were likely to play a regulatory role. Finally the motifs over-represented in sorghum DREBs in comparison to their rice orthologs were identified. Results indicated over-representation of motifs pertaining to calcium, light, sugar, and hormone signaling in the DREB promoters. The co-localized motifs in DREB promoters were mainly those involved in abscisic acid-, light- and calcium-mediated regulation. These motifs along with others pertaining to ethylene signaling were over-represented in sorghum DREB promoters as compared to their orthologs from rice and could possibly contribute to its drought tolerance. Besides calcium, an integration of abscisic acid, ethylene, auxin and methyl jasmonate signaling was probably involved in regulating expression of the drought response through DREB transcription factors.
Transcription factors belonging to the AP2-ERF family bind to a DRE/CRT element (core sequence A/GCCGAC) present in the promoters of genes involved in growth and development as well as in stress responses. About 170 genes in rice (Oryza sativa) and 178 genes in sorghum (Sorghum bicolor) (www.grassius.org) are known to belong to this family of transcription factors. Phylogenetic analysis of these genes reveals the existence of several groups, each showing structural and functional similarity.Citation1,Citation2 The cold-inducible DREB1 genes are classified into Group IIIc and consist of 11 genes in rice, while the drought-inducible DREB2 genes are classified into Group IV and consist of 6 genes in rice.Citation2 The DREB proteins are characterized by the presence of several conserved domains besides the AP2 domain, which differ in the DREB1 and DREB2 proteins.Citation2 The DREB proteins bind to a cis-element called the dehydration responsive element (DRE) or C-repeat (CRT) consisting of a core sequence A/GCCGAC, though their affinities to variation in this sequence differ.Citation3,Citation4 This leads to activation of common (dehydrins, LEA proteins, transcription factors, protein kinases) as well as distinct (starch-degrading enzymes by DREB1; heat shock proteins by DREB2) downstream genes by the DREB1 and DREB2 proteins.Citation5 Many cold- or drought-responsive genes are also known to have additional regulatory motifs like abscisic acid response elements (ABREs) in their promoters, suggesting cross-talk between different regulatory pathways under stress conditions.Citation6
Though the regulation of downstream genes by DREB proteins has been fairly well studied, the regulation of DREB genes themselves, under cold or drought stress conditions, is little understood. A transcription factor ICE1 (inducer of CBF expression 1), which codes for a MYC type of transcription factor was identified as a regulator of DREB1A expression,Citation7 while a drought-inducible DREB1D gene was identified in Arabidopsis that was regulated by the ABA-signaling pathway.Citation8
In order to understand the possible mechanisms involved in regulation of DREB gene expression, we carried out an in silico analysis of conserved motifs in the promoters of DREB genes from rice and sorghum. The two species show about 60% synteny and the exon size distribution and intron positions of orthologous sorghum and rice genes agree closely.Citation9 However the two species differ in the extent of their ability to tolerate drought, where sorghum exhibits greater drought tolerance than rice. Over-represented motifs in DREB1 and DREB2 promoters were identified by scoring the frequency of their occurrence in the DREB promoters as compared to all the promoters in rice and sorghum genomes. Subsequently the common occurrences and order of these motifs in the promoters of orthologous DREB genes of sorghum and rice were studied to identify the motifs that were most likely to play a role in regulating the expression of DREB genes. We show here that abscisic acid signals, probably integrated with light and calcium signals, play a role in regulating DREB gene expression at the transcriptional level.
Sequences of 17 rice DREB genes identified by Nakano et al.Citation2 and 1,000 nt upstream regions of these genes were downloaded from the Gramene database (http://www.gramene.org). Of the 178 sequences of AP2/ERF family of sorghum transcription factors available at Gramene, 15 DREB genes were identified on the basis of conserved sequencesCitation2 lying outside the AP2 domain and 1,000 nt upstream sequences were downloaded as for rice. A phylogenetic tree for the DREB protein sequences of rice and sorghum was constructed using the maximum parsimony method using 1,000 bootstrap replicates (MEGA version 4,Citation10).
The rice promoter database comprising of promoters for all the genes in the rice genome (Rice Genome Annotation: http://rice.plantbiology.msu.edu/) was used. Chromosome coordinates of all the genes in sorghum genome (Sorbi1_assembly_scaffolds.fasta.gz) were used to identify and download the 1,000 nt upstream regions. The promoter sequences of all DREB genes (32) were compared with the promoter sequences for all the remaining genes in the rice and sorghum genomes (88,659 genes). An in-house developed PERL program was used for identifying motifs from the PLACE database and other collected motifs.Citation11 Significantly over-represented individual motifs were identified on the basis of the number of genes showing greater than T (where T = 1, 2......20) occurrences of the motif as compared to the number of genes showing less than T occurrences of the same motif within the clusters of DREB genes and the remaining genes of the genome, respectively. Additionally, only those motifs that were represented in more than 50% of the DREB genes were considered. Motifs showing significant over-representation (p ≤ 0.03) were identified using one sample test for binomial proportion.Citation12 The same PERL program was also used for identifying motifs that were over-represented in sorghum DREB promoters as compared to the rice DREB promoters. Motifs present in at least 50% of the sorghum DREB promoters and which showed significance values ≤0.05 using Fischer's one-sided exact test were considered as over-represented.
Rice orthologs for each of the 15 sorghum DREB genes were identified using the ‘Orthologues’ function in gene-based displays available at Gramene (). Of the 17 rice DREB genes only 13 were orthologous to the 15 sorghum DREB genes and promoters of only these 13 rice genes were used for analysis. Motifs common to both rice and sorghum DREB orthologs, motifs common to only DREB1 orthologs and motifs present in only DREB2 orthologs were identified from the promoter regions of the orthologous gene pairs using standard string matching.
The 1,000 nt promoter sequences of the DREB orthologs from sorghum and rice were aligned using CLUSTALW (http://www.ebi.ac.uk). A phylogenetic tree was constructed for the promoter sequences using maximum parsimony method with 1,000 bootstrap replicates (MEGA version 4,Citation10). Co-localized motifs were identified in each orthologous DREB promoter pair (both strands) using string matching. Motifs within 100 nt of each other in an ortholog pair, which were similarly ordered on the promoter sequences, were considered as co-localized.
The DREB genes identified from rice and sorghum are shown in . The similarity in protein sequences of these 32 DREB genes (DREB1 and 2) was studied by aligning the sequences and constructing a dendrogram. DREB1 and DREB2 proteins formed two major branches, with the DREB2 proteins separating on a single branch and most of the DREB1 genes into another branch (). One DREB1 protein from rice, Os06g07030 (identified as DREB1D) however was seen to be grouped on the same branch as the DREB2 proteins.
Using an in-house developed PERL program, the motifs over-represented in the pooled DREB1 and DREB2 promoters of sorghum and rice as compared to the rest of the respective genomes were identified. Only those motifs occurring in at least 50% of the DREB promoters and which showed a p value ≤0.03 were considered (one-sample test for binomial proportion) (). Three motifs were over-represented in both DREB1 and DREB2 genes, which corresponded to the cis-elements that play an important role in Ca+2/Calmodulin signaling (one) and binding of ABA-induced transcription factors (two). The ABREs (ABA response elements) were represented at least once in DREB 1 promoters and at least twice in DREB2 promoters.
The DREB1 promoters alone showed over-representation of 17 other motifs, many of which were involved in hormone signaling (auxin, GA, ethylene, ABA, MeJA). A few motifs were coupled to light-regulated responses, sugar signaling and anaerobiosis. Two motifs represented binding sites for nuclear factors. On the other hand only two motifs were overrepresented in the DREB2 promoters, one of which was an ABRE and the other one was involved in light-regulated responses.
Fifteen orthologous pairs for sorghum and rice DREB genes were identified () using the Orthologues function in the gene-based displays available at the Gramene database (http://www.gramene.org). The 1,000 nucleotide upstream promoter sequences of these gene pairs were compared for detecting common occurrences. Both + and − strands of an orthologous pair of sorghum and rice promoters were screened for motifs that were shown to be over-represented in DREBs (cf. ). Eleven motifs were common to orthologous pairs of both DREB1 and DREB2 promoters, which included motifs involved in hormone and stress signaling and two motifs involved in light-regulated responses ().
Sequence similarity between the promoters of orthologs was studied by aligning the sequences and constructing a dendrogram, using MEGA version 4 (maximum parsimony with 1,000 bootstrap replicates). In this tree, there was no clear separation of the DREB1 and DREB2 promoters into separate groups ().
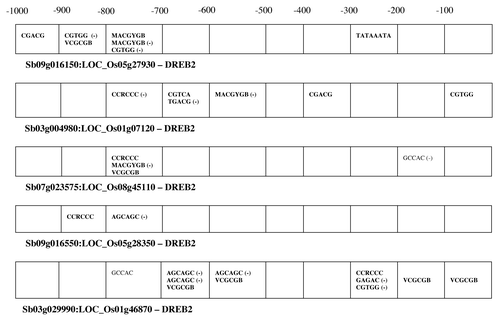
The aligned promoter sequences of each orthologous pair were also screened for co-localized motifs using string matching. All, except one, of the orthologous pairs of DREB promoters (Sb07g02510:Os08g43200) showed at least one of the two motifs representing ABREs namely MACGYGB and CGTGG (). Twelve of the fifteen orthologous pairs showed co-localization of the motif VCGCB, which plays a role in the regulation of gene expression in response to Ca+2/Calmodulin signal. The orthologous pair (Sb07g02510:Os08g43200) that lacked the ABRE motifs, showed three co-localized motifs related to sugar signaling. Besides ABREs, most of the DREB promoters showed co-localized motifs for light regulated response (CCRCCC), MeJA responsive motifs (CGTCA) and auxin response (GAGAC and TGACG). Two DREB 2 promoters showed the presence of a regulatory motif involved in anaerobiosis signaling.
These co-localized motifs were ordered on each of the orthologous pairs of chromosomes to study their location with respect to each other (). The position of the motifs of both + and − strands was taken into consideration. Most of the co-localized motifs were located beyond 400 nucleotides from the start codon (ATG) of DREB1 genes. Multiple motifs for ABREs and Ca+2/Calmodulin signaling were observed to lie near each other. Three of the DREB 1 promoters also showed presence of the motif for light regulated response in the same region. The location of co-localized motifs was more diverse in DREB 2 promoters and one of them (Sb03g029990:Os01g46870) showed an aggregation of motifs for light, Ca+2/Calmodulin, ABA response and MeJA response within the 300 nucleotides upstream region. This promoter also showed the presence of three motifs for anaerobiosis signaling. A TATAAATA box motif was co-localized in the −200 to −300 upstream region of one orthologous pair (Sb09g016150: Os05g27930) and, moreover this promoter lacked the regulatory motif for light signaling, which was present in all other DREB 2 promoters.
Using the same PERL program described earlier, seven motifs were identified as over-represented in all the sorghum DREB promoters (15) as compared to their representation in rice DREB promoters (17). These included the light-inducible GCCAC motif that was present at least twice in all the sorghum DREB1 and four out of five DREB2 promoters respectively (). The VCGCGB motif that was co-localized in several orthologous pairs of sorghum and rice DREB promoters, showed over-representation in the sorghum DREB promoters. Three motifs pertaining to low temperature, pathogen and ethylene signaling were also over-represented in the sorghum DREB promoters.
Availability of the sorghum and rice genome sequences has made it possible to compare the regulatory motifs over-represented in promoters of specific genes as compared to the rest of the promoters in the entire genomes. DREB genes are known to be a distinct sub-group of the AP2-ERF family of transcription factors that regulate the expression of a large number of downstream genes involved in drought, salt or low temperature responses of a plant. However, little is known about the factors regulating expression of the DREB genes themselves.
Promoter regions of the 21 DREB1 and 11 DREB2 genes from sorghum and rice used in this study showed over-representation of several motifs as compared to all the other promoters in their genomes. Many of the DREB genes showed the presence of the VCGCGB and MACGYGB motifs. These motifs are known to function as Ca2+ and ABA responsive cis-elements in plants to which the calmodulin-binding transcription activators (CAMTAs) bind and are present in the promoters of several cold-regulated genes.Citation13 Another ABRE motif, CGTGG was also over-represented in the DREB gene promoters of sorghum and rice. Additionally, DREB1 promoters showed over-representation of two more ABRE motifs (ACGTSSSC and CANNTG) and DREB2 promoters showed the presence of one ABRE motif (CATGCA). ABA levels are known to increase in response to abiotic stresses like drought and salinityCitation14 and the presence of so many ABREs in the promoters of DREB genes indicate a role for ABA-signaling in the regulation of DREB gene expression. However ABA is only a sub-component of the drought responseCitation15 and regulation of the drought-responsive DREB genes are likely to involve several additional factors.
Though the dendrograms depicting phylogenetic relationships between DREB proteins showed distinct branches of DREB1 and DREB2, the corresponding promoter sequences did not separate the two types of DREB genes, indicating that the two groups of DREBs probably share regulatory sequences. Using the promoter sequences of orthologous genes of rice and sorghum DREBs, we looked for motifs from among the over-represented motifs that showed common occurrences in the orthologs. Nine motifs representing hormone, light and sugar regulation were common to more than 50% of the identified orthologous pairs of both DREB1 and DREB2 promoters. Further, we reasoned that if the common motifs in the orthologous pairs were co-localized, then such motifs could be considered to be important for transcriptional regulation. It was observed that most of the orthologous DREB promoter pairs showed co-localization of motifs involved in ABA, Ca2+ and light responses indicating that these factors play an important role in regulating DREB genes. Four of the DREB1 orthologous pairs and one DREB 2 orthologous pair also showed co-localized motifs for MeJA signaling. MeJA levels are known to increase in response to abiotic stresses like drought and salinityCitation16 as well as in response to biotic stresses.Citation17 MeJA responsive expression of a rice gene involved in defense responses (OsOPR1) has been seen to be mediated through a bZIP transcription factor,Citation18 which binds to the cis-elements TGACG and ACGT. The presence of one of these motifs, TGACG in DREB genes was suggestive of a role for bZIP factors in regulating MeJA-induced regulation. Some of the DREB promoters showed co-localized motifs for auxin signaling involving auxin response factors (ARFs). In Phaseolus vulgaris, an ABA responsive transcription factor ABI3 was shown to bind to both ABA and auxin response elements.Citation19 Hence cross-talk between ABA and auxin signaling may be involved in regulating the expression of DREB genes.
The dehydration stress response in plants is mediated by both ABA-dependent and ABA-independent pathways and the DREB regulon is known to function in the latter pathway.Citation20 However, over-representation of several ABREs in the promoters of DREB genes suggests that ABA may play an important role in regulating the expression of DREB transcription factors, which then regulate the expression of dehydration response genes in an ABA-independent manner.
Most of the DREB genes also showed the presence of motifs regulating light and sugar responses. Abiotic stresses lead to an arrest of plant growth, which would necessitate regulation of photosynthesis and carbohydrate partitioning.Citation21 The presence of the light and sugar regulatory motifs in DREB promoters indicates an integration of growth responses and stress responses in a plant.
Though sorghum and rice showed a high extent of homology in the DREB genes and their promoters, sorghum is known to express drought tolerance while rice does not. We looked for over-represented motifs in the DREB promoters of sorghum as compared to rice to understand whether expression of the DREB genes differs in these two species. The sorghum DREB promoters showed over-represented motifs for light (GCCAC), Ca2+ and ethylene signaling as compared to the rice promoters. Ethylene induced DNA binding proteins include transcription factors like EIN3 and ethylene responsive element binding proteins that bind to the GCC box present in the promoters of many ethylene responsive genes. In addition, an ethylene induced calmodulin binding protein (EICBP) was identified in Arabidopsis that showed the CG-1 binding motif that is common to CAMTA proteins that bind to the VCGCGB motif involved in Ca2+ calmodulin signaling, as well as to the Ca2+ responsive ABRE motif (MACGYGB). The DREB1 genes of sorghum also showed over-representation of the ICEr2 motif that is known to play a role in expression of the cold-induced DREB1c gene (CBF2) from Arabidopsis.Citation22 Moreover the ICEr2 motif overlaps the VCGCB motif in Arabidopsis. Thus over-represented motifs for Ca2+ and ethylene signaling together with the ABREs may play an important role in integrating the ethylene and ABA signals in sorghum DREB genes and contribute to its better ability to withstand abiotic stress than rice, which requires experimental validation.
Figures and Tables
Figure 1 Dendrogram showing the clustering of DREB 1 (black) and DREB 2 (gray) proteins of sorghum and rice. The dendrogram was constructed using the maximum parsimony algorithm with 1,000 bootstrap replicates (MEGA version 4).
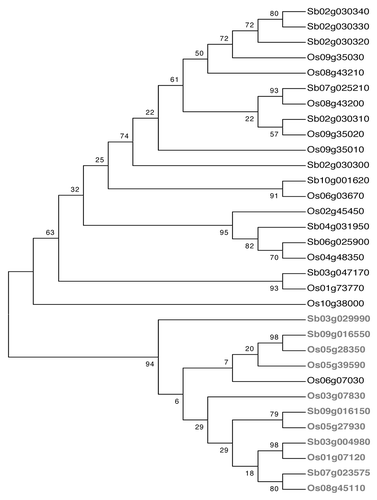
Figure 2 Dendrogram showing the clustering of −1,000 nt DREB 1 (black) and DREB 2 (gray) promoter sequences of sorghum and rice. The dendrogram was constructed using the maximum parsimony algorithm with 1,000 bootstrap replicates (MEGA version 4).
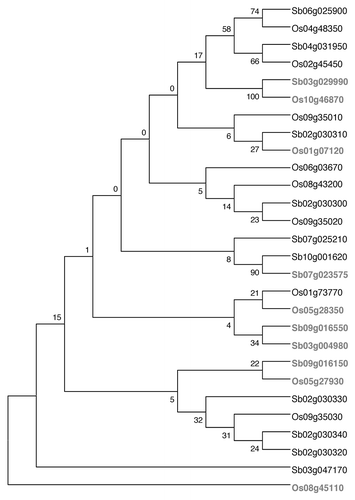
Figure 3 Over-represented motifs that are co-localized on the promoter sequences of orthologous sorghum and rice DREB genes. (−) indicates presence of the motif on the negative strand.
Table 1 DREB genes of rice and sorghum
Table 2 Orthologous pairs of sorghum and rice DREB genes
Table 3 Motifs over-represented in promoters of sorghum and rice DREB1 (21 genes) and DREB2 genes (11 genes)
Table 4 Motifs showing common occurrence in the promoters of 15 orthologous DREB genes of sorghum and rice
Table 5 Motifs co-localized in aligned promoters of DREB1 and DREB2 orthologs from sorghum and rice
Table 6 Motifs over-represented in DREB promoters of sorghum (15) as compared to their occurrence in DREB promoters of rice (17)
Acknowledgements
S.B. and A.S. acknowledge the financial assistance provided by Department of Biotechnology (DBT), Government of India, and also thank Dr. Sharayu Paranjpe, Statistics Department, University of Pune, for advice on statistical methods used. A.L. acknowledges the financial assistance provided by the School of Life Sciences, University of Skövde
References
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 2002; 290:998 - 1009
- Nakano T, Suzuki K, Fujimura T, Shinshi H. Genomewide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 2006; 140:411 - 432
- Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, et al. Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J 2004; 38:982 - 993
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, et al. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 2006; 18:1292 - 1309
- Maruyama K, Takeda M, Kidokoro S, Yamada K, Sakuma Y, Urano K, et al. Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol 2009; 150:1972 - 1980
- Nakashima K, Yamaguchi-Shinozaki K. Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiol Plant 2006; 126:62 - 71
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, et al. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 2003; 17:1043 - 1054
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 2002; 130:639 - 648
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, et al. The Sorghum bicolor genome and the diversification of grasses. Nature 2009; 457:551 - 556
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 2007; 24:1596 - 1599
- Lindlof A, Brautigam M, Chawade A, Olsson O. In silico analysis of promoter regions from cold-induced genes in rice (Oryza sativa L.) and Arabidopsis thaliana reveals the importance of combinatorial control. Bioinformatics 2009; 25:1345 - 1348
- Snedecor GW, Cochran WG. Statistical methods 1994; 8th Edition New Delhi Affiliated East West Press 117 - 120
- Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 2009; l21:972 - 984
- Xiong L, Zhu J-K. Regulation of abscisic acid biosynthesis. Plant Physiol 2003; 133:29 - 36
- Buchanan CD, Lim S, Salzman RA, Kagiampakis I, Morishige DT, Weers BD, et al. Sorghum bicolor's transcriptome response to dehydration, high salinity and ABA. Plant Molec Biol 2005; 58:699 - 720
- Creelman RA, Mullet JE. Jasmonic acid distribution and action in plants: regulation during development and response to biotic and abiotic stress. Proc Natl Acad Sci USA 1995; 92:4114 - 4119
- Turner JG, Ellis C, Devoto A. The jasmonate signal pathway. Plant Cell 2002; 14:153 - 164
- Sobajima H, Tani T, Chujo T, Okada K, Suzuki K, Mori S, et al. Identification of a jasmonic acid -responsive region in the promoter of the rice 12-oxophytodienoic acid reductase 1 gene OsOPR1. Biosci Biotechnol Biochem 2007; 71:3110 - 3115
- Nag R, Maity MK, DasGupta M. Dual DNA Binding property of ABA insensitive 3 like factors targeted to promoters responsive to ABA and auxin. Plant Mol Biol 2005; 59
- Nakashima K, Yusuke I, Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 2009; 149:88 - 95
- Bartels D, Sunkar R. Drought and salt tolerance in plants. Crit Rev Plant Sci 2005; 24:23 - 58
- Zarka DG, Vogel JV, Cook D, Thomashow MF. Cold induction of arabidopsis CBF genes involves multiple ICE promoter elements and a cold regulatory circuit that is desensitized by low temperature. Plant Physiol 2003; 133:910 - 918