Abstract
The WOX (WUSCHEL-RELATED HOMEOBOX) gene family of Arabidopsis comprises fifteen plant-specific transcriptional factors that play important development roles. Genetic, phylogenetic, and genomic analyses suggest that WOX genes generally act non-autonomously to organize stem-cell and initial-cell populations within plant meristems and organ anlagen. Previous cross-complementation analyses indicate that the functional diversification of distinct WOX paralogs may be explained largely by promoter evolution, although paralog-specific protein::protein interactions are also implicated. A recent report described WOX4 function during development of the procambium, which comprises the meristematic tissues of the plant vasculature. Here we show that WOX4 fails to complement PRS1/WOX3 function, when driven from the PRS1/WOX3 native promoter. These data suggest that WOX4 identifies different DNA targets and/or interacting proteins during development of the vasculature procambium than does PRS1/WOX3 during the specification of lateral organ initial cells. The identification of super-compound leaf phenotypes induced by overexpression of the SlWOX4 ortholog in tomato suggests a functional link between vascular patterning and leaf complexity.
WUS1 and 14 WOX (WUSCHEL-RELATED HOMEOBOX) genes are present in Arabidopsis, and five (WUS1, PRS1/WOX3, WOX5, WOX8 and WOX9) are shown genetically to function non-cell-autonomously during the organization of various stem cell populations within plant meristems.Citation1–Citation6 Functional equivalency has been experimentally demonstrated between the shoot stem cell organizer WUS1 and the root stem cell organizer WOX5,Citation7 and also between WUS1 and PRS1, which is required for recruitment of lateral organ founder cells.Citation8 Although these WOX paralogs are normally expressed in distinct stem or initial cell domains, function can be cross-complemented to a high degree when these paralogs are expressed from WOX gene-specific native promoters. Recently we reported that the orthologous genes AtWOX4 and SlWOX4 are expressed in the procambium tissues of stems and leaves in Arabidopsis and tomato, respectively.Citation9 Although WUS1 can fully complement the leaf stipule-less and lateral sepal deletion phenotypes of prs1 mutants when driven by the PRS1 promoter,Citation8 AtWOX4 confers only weak rescue of these prs1 mutant phenotypes. As shown in , although 87% of prs1 mutant plants transformed with pPRS1::AtWOX4 developed flowers with four sepals, only 2% of these developed lateral sepals of normal width (). Moreover, the leaves of these pPRS1::AtWOX4 transformed prs1 mutant plants fail to develop lateral stipules ().
As reported in an initial survey of the WOX gene family in Arabidopsis,Citation2 amino acid sequence similarity among WOX family members is primarily confined to the homeodomain (66 amino acids in WUS1; 65 residues in all other WOX proteins) and the WUS box, a shorter motif of eight amino acids that is conserved in 14 of the 15 WOX proteins and which mediates both repressor and activator functions of the bifunctional WUS protein.Citation10 Whereas WUS1 contains 66% amino acid identity and 80% similarity within the 65 amino acids of the PRS1/WOX3 homeodomain, WOX4 also shares 66% identity but just 76% similarity to PRS1/WOX3. WOX4 is slightly more similar to PRS1/WOX3 within the WUS box, wherein WOX4 shares 7/8 identical and 7/8 similar residues with PRS1/WOX3 and WUS1 contains just 5/8 identical and 6/8 positive amino acids. However, PRS1/WOX3 and WUS1 share three additional predicted protein motifs that are not present in WOX4 (), which may contribute to their differential ability to rescue PRS1/WOX3 function. These include: (1) a seven residue histidine-rich HIS-box located 52 amino acids downstream of the homeodomain in WUS1 and 50 amino acids downstream in PRS1/WOX3; (2) a protein kinase-C phosphorylation site located at position three of the homeodomain in PRS1/WOX3 and WUS1; and (3) an N-myristoylation site located at residues 79–84 after the homeodomain in WUS1 and 81–86 residues downstream in PRS1/WOX3. These data are also consistent with the detailed phylogenetic analysis performed by Vandenbussche et al.Citation11 in which WUS1, WOX5, PRS1/WOX3 and WOX1 comprise a clade that is separate from WOX4 and WOX2. The failure of WOX4 to rescue PRS1/WOX3 function during lateral organ founder cell initialization suggests that the downstream transcriptional targets of the predicted PRS1/WOX3 transcription factor are more shared with WUS1 (and by implication WOX5) than with WOX4, and that meristem and leaf initial cells may be organized by different signaling pathways than are vascular initials.
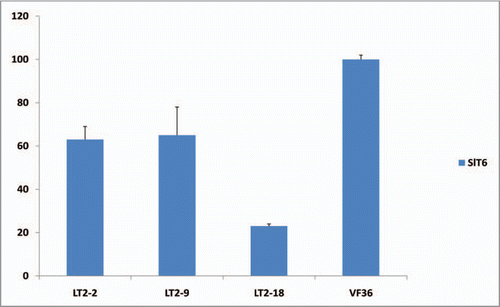
Overexpression of the tomato WOX4 ortholog SlWOX4 alters the vascular development of leaves and stems in tomato.Citation9 Another intriguing phenotype observed in 5S:3SlWOX4 tomato plants is the development of supercompound leaves in which the numbers of primary, intercalary, secondary and tertiary leaflets are all increased (). In addition, supernumery leaflets are produced from the middle of rachis of SlWOX4 overexpressing plants, a phenotype that is not observed in wild type (VF36) leaves (). To investigate the mechanisms underlying the formation of these highly compound leaves, we examined whether the transcript accumulation of the class I KNOX gene SlT6 is altered in SlWOX4 overexpresssing plants. In tomato, overexpression of SlT6 promotes leaf complexity and renders supercompound phenotypes.Citation12,Citation13 Unexpectedly, transcript accumulation of SlT6 in 35S:SlWOX4 plants is downregulated to 23–65% of level observed in wild type plants (). Our data suggest a close connection between vascular patterning and leaf complexity in tomato, and that overexpression of SlWOX4 renders supercompound leaf phenotypes either in a separate pathway than, or downstream of, SlT6. Further characterization of SlWOX4 function in SlT6 mutant plants may help clarify the genetic mechanisms underlying the SlWOX4 supercompound leaf phenotypes.
Figures and Tables
Figure 1 pPRS1::AtWOX4 only partially rescue the phenotype of prs1 mutant. (A) Whole plant phenotype of non-mutant Ler, prs1 mutant, and a transgenic pPRS1::AtWOX4 expressing AtWOX4 coding region under the native PRS1 promoter (2,742 bp) in prs1 mutant.Citation8 (B–D) Stage 12 floral phenotypes of plants shown in (A). Ler (B) flowers have four sepals of normal width and prs1 mutant (C) flowers typically lack lateral sepals. Although four sepals form in pPRS1::AtWOX4 transgenic (D) flowers, the lateral sepals are narrow and the underlying floral organs exposed. (E–G) Transverse sections of shoot apices from vegetative seedlings show development of lateral stipules (arrows) in rosette leaves of Ler, but not in prs1 and pPRS1::AtWOX4 transgenic.

Figure 2 Sepal number but not sepal size in prs1 mutant is rescued by pPRS1::AtWOX4. Total sepal number initiated (A) is reduced in prs1 mutant flowers, but is restored to near normal levels in pPRS1::AtWOX4 transgenic mutant plants. However, sepal size reduced in prs1 mutant relative to Ler floral buds (B) is not complemented by pPRS1::AtWOX4.
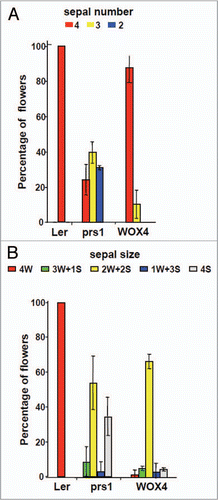
Figure 3 Shared motifs in WUS1, PRS1/WOX3 and WOX4 homologous proteins. Alignment of the full-length amino acid sequences of the WUS1, PRS1/WOX3 and WOX4 proteins. Shared residues found in all three proteins are shown in red; residues shared in two of the three proteins are shown in blue. The WOX homeobox region is bracketed, and comprises 66 amino acids in WUS1 and 65 amino acids in PRS1/WOX3 and WOX4. The HIS-box motifs are boxed in red; the WUS boxes are boxed in blue; the blue asterisks designate shared predicted Protein Kinase-C phosphorylation sites; the red triangles designate predicted shared N-myristoylation sties.
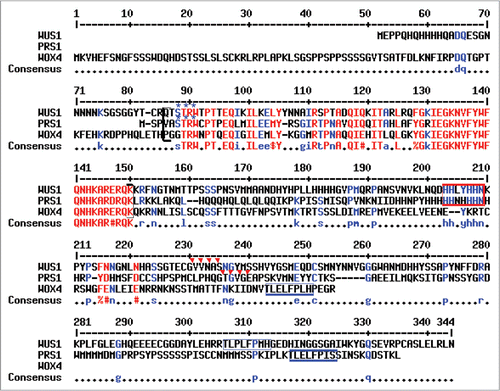
Figure 4 Overexpression of SlWOX4 confers highly compound leaves. (A) Wild type VF36 plant. (B) SlWOX4 overexpressed VF36 at the same stage. (C) Mature leaves from wild type VF36 (left) and SlWOX4 overexpressed VF36 (right). (D) Leaflets (arrows) develop from the middle of rachis (arrowhead) in SlWOX4 overexpressed VF36.

Figure 5 Gene expression levels in the leaves of tomato SlWOX4 overexpressors. Young leaves at the same stages (1 cm long) from three independent transformants (T1–T3) overexpressing SlWOX4 transcripts and from non-transformant VF36 were collected. Quantitative real-time RT-PCR data of SlT6 transcript accumulation is shown relative to the level in the leaves of VF36, which were set to 100%. Three biological replicates were assayed for all samples and normalized to SlACTIN transcript level as described in Ji et al.Citation9
Acknowledgements
This work was supported by the National Science Foundation (grant nos. 517070 and 0649810 to M.J.S.).
Addendum to:
References
- Matsumoto N, Okada K. A homeobox gene PRESSED FLOWER regulates lateral axis-dependent development of Arabidopsis flowers. Genes Dev 2001; 15:3355 - 3364
- Haecker A, Groß-Hardt R, Geiges B, Sarkar A, Breuninger H, Marita Herrmann M, et al. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 2004; 131:657 - 668
- Nardmann J, Ji J, Werr W, Scanlon MJ. The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems. Development 2004; 131:2827 - 2839
- Park SO, Zheng Z, Oppenheimer DG, Hauser BA. The PRETTY FEW SEEDS2 gene encodes an Arabidopsis homeodomain protein that regulates ovule development. Development 2005; 132:841 - 849
- Wu X, Dabi T, Weigel D. Requirement of homeobox gene STIMPY/WOX9 for Arabidopsis meristem growth and maintenance. Curr Biol 2005; 15:436 - 440
- Breuninger H, Rikirsch E, Hermann M, Ueda M, Laux T. Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev Cell 2008; 14:867 - 876
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 2007; 446:811 - 814
- Shimizu R, Ji J, Kelsey E, Schnable PS, Ohtsu K, Scanlon MJ. Tissue-specificity and evolution of meristematic WOX3 function in Arabidopsis. Plant Physiol 2009; 149:841 - 850
- Ji J, Strable J, Shimizu R, Koenig D, Sinha N, Scanlon MJ. WOX4 promotes procambial development. Plant Physiol 2010; 152:1346 - 1356
- Ikeda M, Mitsuda N, Ohme-Tagaki M. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 21:3493 - 3505
- Vandenbussche M, Horstman A, Zethof J, Koes R, Rijpkema AS, Gerats T. Differential recruitment of WOX transcription factors for lateral development and organ fusion in Petunia and Arabidopsis. Plant Cell 2009; 21:2269 - 2283
- Chen JJ, Janssen BJ, Williams A, Sinha N. A gene fusion at a homeobox locus: alternations in leaf shape and implications for morphological evolution. Plant Cell 1997; 9:1289 - 1304
- Janssen BJ, Lund L, Sinha N. Overexpression of a homeobox gene, LeT6, reveals indeterminate features in the tomato compound leaf. Plant Physiol 1998; 117:771 - 786