Abstract
Proper control of stem cell populations is key for the development of all multicellular organisms. In Arabidopsis, stem cells are located primarily in the shoot, root and floral meristems where they undergo complex regulation. The Arabidopsis shoot and root meristems are regulated by the related WUS and WOX5 pathways, respectively. Previous studies established that these pathways share the signal transduction components POLTERGEIST (POL) and PLL1. Our latest study in Plant Cell revealed key roles for acyl modifications and lipid binding in the regulation of these two type 2C protein phosphatases. Specifically, POL and PLL1 were shown to localize to the plasma membrane in a myristioylation- and palmitoylation-dependent manner, POL and PLL1 were shown to bind to membrane lipids, and POL activity was found to be stimulated in vitro by the phospholipid PI(4)P. Here, we will discuss what is currently known in Arabidopsis and other organisms about the mechanisms of palmitoylation and provide additional evidence supporting that POL and PLL1 are palmitoylated, including describing the identification of a putative Arabidopsis palmitoyl transferase as a PLL1 interactor.
The related CLAVATA3 (CLV3)/WUSCHEL (WUS) and CLE40/WOX5 pathways are key for proper regulation of the Arabidopsis shoot and root stem cell populations, respectively.Citation1 The only known signaling intermediates for these pathways are their shared intracellular components, the protein phosphatase type 2C proteins, POLTERGEIST (POL) and PLL1.Citation2–Citation8 Until recently, nothing was known about the mechanisms by which the plasma membrane receptors of these pathways negatively regulate POL and PLL1, nor was anything known about the mechanisms by which POL and PLL1 positively regulate WUS/WOX5 transcription in the nucleus. Our recent study in Plant Cell provides some insights into these questions by revealing that POL and PLL1 are dual acylated plasma membrane proteins that are activated by phospholipids in vitro.Citation9 Significantly, the plasma membrane localization of POL and PLL1 places them in proximity to the transmembrane receptors that negatively regulate their activity.
In our recent study, we found that POL and PLL1 localize to the plasma membrane in a myristoylation- and palmitoylation-dependent manner.Citation9 Furthermore, we observed that these acyl modifications are required for proper POL/PLL1 function in vivo.Citation9 Specifically, our mutational analyses suggested that myristoylation occurs at the second amino acid, a glycine, of each protein following removal of the N-terminal methionine, while palmitoylation occurs at a conserved cysteine residue located nine amino acids carboxyl to the myristoylation site (). Additionally, when both of these modifications were blocked in the PLL1myrmpalm-GFP mutant isoform, we found that the mutated protein functioned in a dominant negative manner in vivo, confirming the importance of dual acylation for POL/PLL1 function. Furthermore, LC/MS/MS analysis of purified, trypsinized, wheat germ-expressed POL revealed that the N-terminal tryptic fragment, MGNGTSR, was not detected. Instead, we identified an 801 dalton myristoyl-GNGTSR fragment, confirming direct myristoylation of POL. Additionally, through MS/MS analysis we were able to identify a mass species of 1,400 daltons which corresponds to the palmitoylated version of the POL tryptic fragment, VVGCFVPSNDK. Unfortunately, we were not able to obtain LC/MS/MS data for this fragment so this assignment, and thus direct evidence of POL/PLL1 palmitoylation, has not been confirmed.
To further support that PLL1 is palmitoylated in planta, we have performed additional confocal analyses on the various Arabidopsis GFP fusion lines from the Plant Cell study following treatment with the palmitoyltransferase inhibitor, 2-bromopalmitate.Citation9,Citation10 Previously, it was shown that treatment of Arabidopsis roots with this palmitate analog results in a phenotype similar to that of the Arabidopsis palmitoyltransferase mutant, TIP1.Citation11 This result shows that Arabidopsis roots are permeable to this inhibitor and that this inhibitor is able to negatively regulate at least some portion of the Arabidopsis palmitoyltransferases. As plasma membrane localization of PLL1 is dependent upon myristoylation and palmitoylation, we hypothesized that if we treated the roots of plants expressing the partially mislocalized unmyristoylatable PLL1myrm-GFP with 2-bromopalmitate, we should also block palmitoylation of this protein. Thus, chemically blocking palmitoylation of the PLL1myrm isoform, which also cannot be myristoylated, should result in a localization pattern similar to that of the PLL1myrmpalm mutant isoform, which cannot be myristoylated or palmitoylated.Citation9 When four day old seedlings expressing PLL1myrm-GFP were treated with 2-bromopalmitate it was clear that this treatment resulted in a dramatic change in the localization pattern of the fusion protein (). Specifically, the protein no longer localized predominantly to the plasma membrane but was instead found in the nucleus and cytoplasm similar to the non-acylatable PLL1myrmpalm-GFP. The effect of 2-bromopalmitate on PLL1-GFP localization was also examined. Here, there were only minor changes in the localization pattern of the fusion protein with more of the protein present in the cytoplasm in the treated sample than the untreated, which mimics the localization seen in the PLL1palm-GFP mutant. Finally, when plants expressing PLL1palm-GFP were treated with 2-bromopalmitate there was little to no change in the localization of the fusion protein. This result is what we would expect as this version of the protein is already unable to undergo palmitoylation and thus treatment with 2-bromopalmitate should have no effect on its overall acylation. Taken together, these results further support that PLL1 undergoes palmitoylation at its 11th amino acid, a cysteine.
In the last eight years, two classes of proteins have been identified as palmitoyltransferases; these are the N-palmitoyltransferases and the S-acyltransferases. The first class, the N-palmitoyltransferases are enzymes that share homology with the O-acyltransferases.Citation12 These palmitoyltransferases are thought to catalyze the addition of palmitoyl to an N-terminal cysteine residue through an amide linkage. The founding member of this family is the Drosophila protein Skinny hedgehog.Citation13 While genetic studies strongly suggest that Skinny Hedgehog and other related proteins are N-palmitoyltransferases there currently is no direct biochemical evidence.Citation13,Citation14
The second class of palmitoyltransferases are the S-acyltransferases. These proteins are transmembrane thiol-directed protein acyltransferases (PATs). Specifically, these enzymes catalyze the attachment of palmitoyl (and sometimes other acyl groups) to specific cysteines within the target proteins through a thioester bond.Citation15 The founding members of this family are the S. cerevisiae proteins, Akr1p and Efr2p, which were the first proteins shown to have palmitoyltransferase activity.Citation12,Citation16,Citation17 All members of this class of palmitoyltransferases contain a cysteine rich DHHC domain thought to be responsible for catalyzing the palmitoyl-transferase activity.Citation15 All DHHC proteins also contain four transmembrane domains with the DHHC domain falling between the second and third transmembrane domains. Finally, there are two other motifs that are highly conserved among DHHC proteins, the DPG motif and the TTxE motif. Outside of these conserved regions the DHHC proteins are highly variable containing numerous other known and unknown protein domains. It is currently thought that all DHHC domain containing proteins may function as S-palmitoyltransferases.Citation15,Citation18
As the POL and PLL1 palmitoylation attachment sites are within the proteins, they undergo palmitoylation through S-acylation. The most obvious candidates then to acylate these phosphatases are the Arabidopsis DHHC-containing proteins.Citation9 In Arabidopsis, twenty-three DHHC-containing proteins have been identified through sequence analysis but only one of these has been studied extensively, TIP1 (At5g20350).Citation11,Citation19 Prior studies have revealed that TIP1 is a bona fide palmitoyltransferase, is involved in a number of processes including root hair and pollen tube growth, and complements the yeast S-acyl transferase mutant akr1Δ.Citation11,Citation20
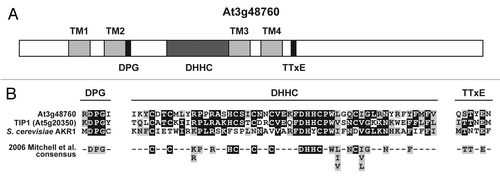
Interestingly, in a screen for proteins interacting with the N-terminal region of PLL1, we have identified a potential Arabidopsis DHHC palmitoyltransferase, At3g48760. Through sequence analysis we have confirmed that At3g48760 contains all of the domains conserved in DHHC palmitoyltransferases (). Furthermore, when the DHHC domain and the DPG and TTxE motifs of At3g48760 are aligned with the corresponding regions of the Arabidopsis palmitoyltransferase, TIP1, and the S. cerevisiae palmitoyltransferase, AKR1, it is clear that the key residues are highly conserved ().Citation15 In summary, the additional confocal studies presented here, further support that POL and PLL1 are palmitoylated in vivo. Additionally, we have found that POL and PLL1 palmitoylation may be controlled in part by the At3g48760 gene product, however further analysis is required to confirm this function.
Figures and Tables
Figure 1 PLL1myrm-GFP is mis-localized following treatment with the palmitoylation inhibitor 2-bromopalmitate. (A) The amino acid sequences of the putative N-myristoylation and palmitoylation signal peptides of POL and PLL1. The glycines and cysteines where the acyl modifications, myristoyl and palmitoyl, are attached, respectively, are shaded in gray. (B) GFP fusion protein localization with or without treatment with the palmitoylation inhibitor, 2-bromopalmitate.Citation10 The PLL1-GFP, PLL1myrm-GFP and PLL1palm-GFP lines are as described.Citation9 Plants were treated for 6 hours with 100 µM 2-bromopalmitate (Sigma) or solvent control as described.Citation11 Confocal images were obtained as describedCitation9 with the following modifications, staining was performed using 10 µg/mL propidium iodide in the treatment solutions and the samples were mounted on the slides in the treatment solutions. While all samples showed minor changes in cellular morphology in response to treatment with 2-bromopalmitate, only the localization of the PLL1myrm-GFP fusion protein was dramatically altered following treatment.
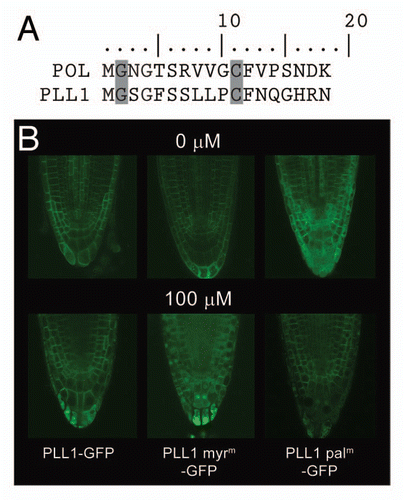
Figure 2 A putative Arabidopsis palmitoyltransferase (S-acyltransferase) interacts with the N-terminus of PLL1. (A) The CytoTrap yeast two hybrid system (Stratagene) was used to screen a flower cDNA library (generated at DNA Technologies, Inc., Gaithersburg, MD) in the vector pMYR (Stratagene) for interactions with the N-terminal 258 amino acids of PLL1. The bait construct was generated by cloning the appropriate region of PLL1 (amplified with the primers GCC CGG GCG ACG TCG ACG ATG GGA AGT GGA TTC TCC TCC and AAT TAA CCG CGG CGG CCG CGA TCA TGC CAT AGC TTC AAC) into the SalI and NotI sites of pSOS (Stratagene). The bait construct was then transformed into yeast strain cdc25H(a) and positive clones were identified following mating with the meristem yeast two hybrid library which was in the strain cdc25H(α).Citation21 Sequence analysis revealed that two of the thirteen N-terminal PLL1 interacting clones encoded At3g48760. At3g48760 is predicted to contain four transmembrane domains (light grey), a DPG (aspartate-proline-glycine) motif (black), a TTxE (threonine-threonine-x-glutamate) motif (black) and a DHHC (aspartate-histidine-histidine-cysteine) domain (medium grey). (B) At3g48760 contains the key residues required for the palmitoyltransferase activity of other members of the DHHC family.Citation15 The DHHC domain and DPG and TTxE motifs from At3g48760 are shown aligned with the corresponding domains of the Arabidopsis palmitoyltransferase, TIP1 (At5g020350) and the S. cerevisiae palmitoyltransferase, AKR1.Citation11,Citation17 The consensus sequences at the bottom are from Mitchell et al. 2006.Citation15
Acknowledgements
This work was supported by National Institutes of Health grant (R01GM62962) to S.E.C.
Addendum to:
References
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 2007; 446:811 - 814
- Gagne JM, Clark SE. The Protein Phosphatases POL and PLL1 are Signaling Intermediates for Multiple Pathways in Arabidopsis. Plant Signal Behav 2007; 2:245 - 246
- Gagne JM, Song SK, Clark SE. POLTERGEIST and PLL1 are required for stem cell function with potential roles in cell asymmetry and auxin signaling. Commun Integr Biol 2008; 1:53 - 55
- Song SK, Clark SE. POL and related phosphatases are dosage-sensitive regulators of meristem and organ development in Arabidopsis. Dev Biol 2005; 285:272 - 284
- Song SK, Hofhuis H, Lee MM, Clark SE. Key divisions in the early arabidopsis embryo require POL and PLL1 phosphatases to establish the root stem cell organizer and vascular axis. Dev Cell 2008; 15:98 - 109
- Song SK, Lee MM, Clark SE. POL and PLL1 phosphatases are CLAVATA1 signaling intermediates required for Arabidopsis shoot and floral stem cells. Development 2006; 133:4691 - 4698
- Yu LP, Miller AK, Clark SE. POLTERGEIST encodes a protein phosphatase 2C that regulates CLAVATA pathways controlling stem cell identity at Arabidopsis shoot and flower meristems. Curr Biol 2003; 13:179 - 188
- Yu LP, Simon EJ, Trotochaud AE, Clark SE. POLTERGEIST functions to regulate meristem development downstream of the CLAVATA loci. Development 2000; 127:1661 - 1670
- Gagne JM, Clark SE. The Arabidopsis stem cell factor POLTERGEIST is membrane localized and phospholipid stimulated. Plant Cell 2010; 22:729 - 743
- Webb Y, Hermida-Matsumoto L, Resh MD. Inhibition of protein palmitoylation, raft localization and T cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J Biol Chem 2000; 275:261 - 270
- Hemsley PA, Kemp AC, Grierson CS. The TIP GROWTH DEFECTIVE1 S-acyl transferase regulates plant cell growth in Arabidopsis. Plant Cell 2005; 17:2554 - 2563
- Linder ME, Deschenes RJ. New insights into the mechanisms of protein palmitoylation. Biochemistry 2003; 42:4311 - 4320
- Chamoun Z, Mann RK, Nellen D, von Kessler DP, Bellotto M, Beachy PA, et al. Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science 2001; 293:2080 - 2084
- Zhai L, Chaturvedi D, Cumberledge S. Drosophila wnt-1 undergoes a hydrophobic modification and is targeted to lipid rafts, a process that requires porcupine. J Biol Chem 2004; 279:33220 - 33227
- Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J Lipid Res 2006; 47:1118 - 1127
- Lobo S, Greentree WK, Linder ME, Deschenes RJ. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J Biol Chem 2002; 277:41268 - 41273
- Roth AF, Feng Y, Chen L, Davis NG. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J Cell Biol 2002; 159:23 - 28
- Dietrich LE, Ungermann C. On the mechanism of protein palmitoylation. EMBO Rep 2004; 5:1053 - 1057
- Hemsley PA. Protein S-acylation in plants. Mol Membr Biol 2009; 26:114 - 125
- Schiefelbein J, Galway M, Masucci J, Ford S. Pollen tube and root-hair tip growth is disrupted in a mutant of Arabidopsis thaliana. Plant Physiol 1993; 103:979 - 985
- Soellick TR, Uhrig JF. Development of an optimized interaction-mating protocol for large-scale yeast two-hybrid analyses. Genome Biol 2001; 2:52 - 57