Abstract
Gibberellins (GAs) are phytohormones that regulate growth and development throughout the life cycle of plants. Negative feedback contributes to homeostasis of GA levels. DELLA proteins are involved in this process. Since DELLA proteins do not have apparent DNA binding motifs, other DNA binding proteins might act as a mediator downstream of DELLA proteins in the GA feedback regulation. In this review, we highlight the mechanisms of GA feedback regulation, specifically the differential regulation of GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox) by transcription factors. RSG (REPRESSION OF SHOOT GROWTH) is a tobacco (Nicotiana tabacum) transcriptional activator with a basic leucine zipper domain that controls the levels of endogenous GAs through the regulation of GA biosynthesis genes. Recently we reported that RSG not only regulates the expression of ent-kaurene oxidase gene but is also involved in the negative feedback of NtGA20ox1 by GAs. RSG plays a role in the homeostasis of GAs through direct binding to the NtGA20ox1 promoter triggered by a decrease in GA levels in the cell. Furthermore, decreases in GA levels promote modifications of active histone marks on the NtGA20ox1 promoter. We have developed a hypothetical model to explain how RSG regulates dual target genes via epigenetic regulation.
GA Homeostasis and GA Signaling
Gibberellins (GAs) are tetracyclic diterpenoid phytohormones that regulate many aspects of plant development, including seed germination, leaf expansion, stem elongation, flowering and flower development.Citation1–Citation3 GA biosynthetic enzymes have been characterized by ways of genetics, molecular biology and biochemistry. In Arabidopsis six GA biosynthetic enzymes have been identified principally through the studies of GA deficient mutants.Citation4 Some of the GA biosynthetic genes contribute to GA homeostasis in plants. For example, the expression of AtGA20ox1, AtGA20ox2, AtGA20ox3 and AtGA3ox1 is induced under GA-deficient conditions, but they are downregulated by application of exogenous GAs.Citation5–Citation7 GA feedback regulation has been shown to depend on GA signaling components, including DELLA proteins, SPINDLY, the F-box proteins SLEEPY1/GID2 and the GA receptors AtGID1/GID1,Citation8–Citation11 with DELLA proteins and SPINDLY being negative regulators, while others are positive regulators in the GA response pathway. Under GA deficient conditions, DELLA proteins accumulate in the nucleus and the transcripts of GA20ox and GA3ox increase concomitantly. Zentella R et al. have identified 14 early GA responsive genes as targets of DELLA protein,Citation12 including AtGA20ox2 and AtGA3ox1. DELLA proteins activate the expression of these genes, but no evidence has been shown that DELLA proteins bind to the AtGA20ox2 and AtGA3ox1 promoter directly. Because DELLA proteins do not have apparent DNA binding motif, it is possible that some other unknown DNA-binding proteins are involved in the direct regulation of these gene along with DELLA protein or they may function as downstream components of DELLA protein. Recent studies revealed that DELLA proteins interact with PHYTOCHROME INTERACTING FACTORS (PIFs), PIF3 and PIF4.Citation13,Citation14 More recent work has shown that DELLA proteins also interact with other PIFs family proteins,Citation15 which suggests that any of these PIF proteins might be involved in the feedback regulation of GA biosynthetic genes.
GA Feedback Regulation by Transcription Factors
Recently transcription factors and cis-elements that are involved in GA feedback regulation have been identified. Overexpression of AGF1, an AT-hook motif protein in Arabidopsis, enhanced upregulation of AtGA3ox1 in response to decrease of GAs, but did not affect the expression of the AtGA20ox genes. AT-hook motif proteins bind to the minor groove of the AT-rich sequence and regulate gene expression with other transcription factors. AGF1 binds to GNFEI, which is a 43 bp cis-element located in the AtGA3ox1 promoter responsible for GA negative feedback regulation. The GNFEI sequence was not found in the promoter region of AtGA20ox1–3. This suggests that the feedback regulation of the AtGA3ox1 through AGF1 is rather specific.Citation6 Since AGF1 is necessary but not sufficient to function alone for the feedback upregulation of AtGA3ox1, it is possible that another transcriptional activator is required in this regulation.
YABBBY1 (YAB1) may be a mediator of GA homeostasis that functions downstream of DELLA proteins in rice, because its expression is DELLA-dependent. Overexpression of YAB1 resulted in a semi-dwarf phenotype with decreased OsGA3ox2 transcript levels, whereas co-suppression of YAB1 in transgenic plants induced expression of OsGA3ox2. YAB1 binds to the GA responsive element (GARE) located in the GA3ox2 promoter, indicating that YAB1 regulates OsGA3ox2 directly.Citation16 The GARE sequence does not exist in the OsGA20ox2 promoter.
RSG is a tobacco transcriptional activator with a basic leucine zipper (bZIP) domain that is involved in the regulation of endogenous amounts of GAs.Citation17 A dominant negative form of RSG represses the expression of the ent-kaurene oxidase (KO) gene in transformed tobacco plants, resulting in a reduction of the endogenous amounts of GAs and inhibition of stem cell elongation, leading to a dwarf phenotype. Interestingly, in these transgenic plants, while the transcript levels of NtGA3ox increased as a response to a reduction of GA levels, the expression levels of NtGA20ox1 were not affected. This indicates that the dominant negative form of RSG inhibited only the feedback regulation of NtGA20ox1, but not that of NtGA3ox. Recently we identified an RSG binding sequence (rbe) in the promoter region of NtGA20ox1. A mutation in the RSG-binding sequence abolished the feedback regulation of NtGA20ox1 by GAs, indicating that the rbe sequence is indispensable for the regulation by RSG. RSG binds to this sequence of the NtGA20ox1 promoter in vivo in response to a decrease in GA levels, and the binding is abolished by exogenous GA treatments.Citation18 Therefore, RSG not only controls the expression of NtKOCitation17 but also the feedback regulation of NtGA20ox1.Citation18 RSG is a candidate as a downstream factor of DELLA proteins, but no direct interactions between DELLA proteins and RSG have been observed. RSG could be function in GA feedback regulation through a DELLA independent pathway. Since these transcription factors regulate GA20ox or GA3ox independently, it indicates that the mechanisms of GA feedback regulation are fundamentally different between GA20ox and GA3ox.
RSG Regulates Two GA Biosynthetic Genes Via Different Mechanisms
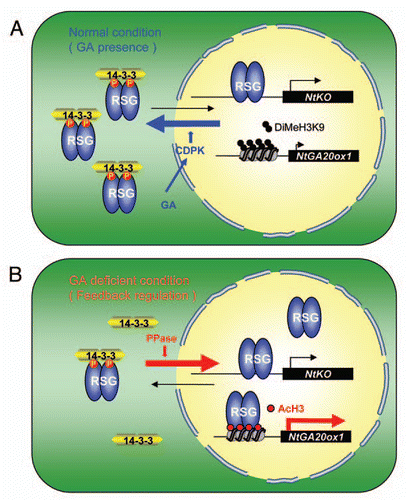
The intracellular localization of RSG is dependent on the levels of GAs in the cell. When the levels of GAs are low, RSG is translocated into the nucleus, whereas GAs treatments reverse this nuclear accumulation.Citation19 The function of RSG is negatively regulated by 14-3-3 proteinsCitation20 which sequester RSG into the cytoplasm by binding to them, and thus prevent RSG function in the nucleus.Citation19,Citation20 The GA dependent cytoplasmic migration of RSG requires the phosphorylation of Ser-114 of RSG by Ser/Thr kinase and 14-3-3 binding.Citation19 NtCDPK1—a Ca2+-dependent protein kinase—was identified as an RSG kinase, in which the variable N-terminal domain is necessary for the recognition of RSG.Citation21 Overexpression of NtCDPK1 inhibits the feedback regulation of NtGA20ox1. Under GA deficient conditions, RSG accumulates in the nucleus, and induce the expression of NtGA20ox1 but not that of NtKO.Citation19 Chromatin immunoprecipitation (ChIP) assays suggested that the binding of RSG to the NtGA20ox1 promoter depends on the endogenous GA levels, but that RSG binding to the NtKO promoter is independent of GA concentrations.Citation18 This indicates different mechanisms for transcriptional regulation of NtKO and NtGA20ox1.
We have developed a hypothetical model for the differential regulation mechanisms of these target genes (). In this model, GA feedback regulation of NtGA20ox1 is controlled by the intracellular localization of RSG and epigenetic regulation by histone modification on the NtGA20ox1 promoter. RSG was shown to bind to the NtGA20ox1 promoter by an in vitro assay. But ChIP assays showed that binding of RSG to the NtGA20ox1 promoter in vivo is limited under GA-deficient conditions, and under normal (GA-sufficient) conditions RSG cannot bind to the NtGA20ox1 promoter.Citation18 The accessibility of transcription factors to the promoter of target genes depends on the modification of the DNA and/or histones via epigenetic regulation. Acetylation of the N-terminal tail of histone H3 generally correlates with transcriptional activity, whereas methylation of histone H3 Lys9 is known to be a transcriptional repressive mark. Under GA deficient condition, there is an increase in the acetylation of histone-H3 and a remarkable decrease in the methylation of histone H3 Lys9 on the region of the NtGA20ox1 promoter containing RSG-binding site, as detected by ChIP assay. These modifications of histone-H3 may serve to control the accessibility of RSG to the NtGA20ox1 promoter,Citation18 suggesting that the endogenous GA levels regulate the chromatin structures around the NtGA20ox1 promoter region. Arabidopsis PKL (PICKLE) could also be involved in the feedback regulation of GA20ox by GAs. PKL codes for a putative CHD3 chromatin remodeling factor and promotes trimethylation or dimethylation of histone H3 Lysine 27 and H3 Lysine 9, contributing to the repression of the target genes by histone H3.Citation22,Citation23 PKL also mediates some aspects of GA responsiveness in the mature plant. Mutations in the PKL gene result in the over-accumulation of bioactive GAs, however, the transcript levels of both AtGA3ox1 and AtGA20ox1 were reduced in pkl mutants, suggesting that PKL is not involved in the feedback regulation of AtGA3ox1 and AtGA20ox1.Citation24 In contrast, the expression of AtGA20ox2 was induced in pkl mutants,Citation23 suggesting that PKL is involved in the repression of AtGA20ox2 expression. PKL and GAs were shown to act concomitantly to repress the expression of embryonic traits in seedlings. In addition, PKL and GAs act synergistically to repress the expression of the same target genes but through separate pathways.Citation23 Similar to the pkl mutation, application of GA biosynthesis inhibitor, uniconazole-P, promotes the expression of AtGA20ox2. The rbe-like sequence was also found in the AtGA20ox2 promoter and RSG was shown to activate AtGA20ox2 in transient assays. Therefore, the regulation of NtGA20ox1 in tobacco and AtGA20ox2 expression in Arabidopsis appears to be similar. The activation of NtGA20ox1 by RSG in tobacco and the repression of AtGA20ox2 by the chromoatin modifier PKL in Arabidopsis might be closely related in the feedback regulation by GAs, because the RSG-binding to the NtGA20ox1 promoter is affected by histone modification and therefore RSG might be involved in epigenetic regulation through histone modification. Further study will be necessary to reveal how GAs regulates the accessibility of RSG to the NtGA20ox1 promoter via epigenetic control.
Figures and Tables
Figure 1 Model of feedback regulation of NtGA20ox1 through the regulation of histone modification and the intracellular localization of RSG. (A) Under normal condition, RSG is localized in both the cytoplasm and the nucleus. RSG is phosphorylated by NtCDPK1, which is activated by GAs. The phosphorylated RSG is sequestered into the cytoplasm by 14-3-3 proteins. Histone H3 Lys9 on the NtGA20ox1 promoter is dimethylated under these conditions preventing the binding of RSG to the NtGA20ox1 promoter. (B) Under GA-deficient conditions, RSG is dephosphorylated and accumulates in the nucleus. Histone H3 Lys9 on the NtGA20ox1 promoter is acetylated, allowing the binding of RSG with the NtGA20ox1 promoter, to activate transcription of the gene.
References
- Hooley R. Gibberellins: perception, transduction and responses. Plant molecular biology 1994; 26:1529 - 1555
- Richards DE, King KE, Ait-Ali T, Harberd NP. HOW GIBBERELLIN REGULATES PLANT GROWTH AND DEVELOPMENT: A molecular genetic analysis of gibberellin signaling. Ann Rev Plant Physiol Plant Molec Biol 2001; 52:67 - 88
- Thomas SG, Rieu I, Steber CM. Gibberellin metabolism and signaling. Vitamins and hormones 2005; 72:289 - 338
- Yamaguchi S. Gibberellin metabolism and its regulation. Ann Rev Plant Biol 2008; 59:225 - 251
- Chiang HH, Hwang I, Goodman HM. Isolation of the Arabidopsis GA4 locus. Plant Cell 1995; 7:195 - 201
- Matsushita A, Furumoto T, Ishida S, Takahashi Y. AGF1, an AT-hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase. Plant Physiol 2007; 143:1152 - 1162
- Phillips AL, Ward DA, Uknes S, Appleford NE, Lange T, Huttly AK, et al. Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol 1995; 108:1049 - 1057
- Schwechheimer C. Understanding gibberellic acid signaling—are we there yet?. Curr Opin Plant Biol 2008; 11:9 - 15
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 2005; 437:693 - 698
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 2006; 18:3399 - 3414
- Sun TP, Gubler F. Molecular mechanism of gibberellin signaling in plants. Ann Rev Plant Biol 2004; 55:197 - 223
- Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, et al. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 2007; 19:3037 - 3057
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 2008; 451:475 - 479
- de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, et al. A molecular framework for light and gibberellin control of cell elongation. Nature 2008; 451:480 - 484
- Gallego-Bartolome J, Minguet EG, Marin JA, Prat S, Blazquez MA, Alabadi D. Transcriptional diversification and functional conservation between DELLA proteins in Arabidopsis. Mol Biol Evol 2010; 27:1247 - 1256
- Dai M, Hu Y, Zhao Y, Liu H, Zhou DX. A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol 2007; 144:380 - 390
- Fukazawa J, Sakai T, Ishida S, Yamaguchi I, Kamiya Y, Takahashi Y. Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 2000; 12:901 - 915
- Fukazawa J, Nakata M, Ito T, Yamaguchi S, Takahashi Y. The transcription factor RSG regulates negative feedback of NtGA20ox1 encoding GA 20-oxidase. Plant J 2010; 62:1035 - 1045
- Ishida S, Fukazawa J, Yuasa T, Takahashi Y. Involvement of 14-3-3 signaling protein binding in the functional regulation of the transcriptional activator REPRESSION OF SHOOT GROWTH by gibberellins. Plant Cell 2004; 16:2641 - 2651
- Igarashi D, Ishida S, Fukazawa J, Takahashi Y. 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 2001; 13:2483 - 2497
- Ito T, Nakata M, Fukazawa J, Ishida S, Takahashi Y. Alteration of substrate specificity: the variable N-terminal domain of tobacco Ca2+-dependent protein kinase is important for substrate recognition. Plant Cell 2010; 22:1592 - 1604
- Perruc E, Kinoshita N, Lopez-Molina L. The role of chromatin-remodeling factor PKL in balancing osmotic stress responses during Arabidopsis seed germination. Plant J 2007; 52:927 - 936
- Zhang H, Rider SD Jr, Henderson JT, Fountain M, Chuang K, Kandachar V, et al. The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. J Biol Chem 2008; 283:22637 - 22648
- Henderson JT, Li HC, Rider SD, Mordhorst AP, Romero-Severson J, Cheng JC, et al. PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol 2004; 134:995 - 1005