Abstract
Shoot apical meristem is a well organized undifferentiated tissue which produces plant body. CLV3 peptide hormone regulates SAM homeostasis, and is perceived by several receptor complexes, CLV1, CLV2-SOL2/CRN, and RPK2. CLV1 homologues are encoded in various plants genome. However CLV2 and SOL2/CRN homologues are found only in higher plants. Here we show that the RPK2 homologues were found not only in moss, Physcomitrella patens, but also in liverwort, Marchantia polymorpha. Although CLV2-SOL2/CRN might have specific function in SAM homeostasis, CLV1 and RPK2 may regulate various plant physiological events during plant evolution.
In higher plants, the CLV signaling pathway regulates meristem fate in a noncellautonomous manner. CLV3 is one of the 32 members of the CLV3/ESR-related (CLE) gene family in Arabidopsis, and CLV3 functions as a dodecapeptideCitation1,Citation2 that functions as an intercellular signaling molecule to repress the expression of the homeobox transcription factor WUS in the organization center. In this signaling pathway, receptor complexes CLV1 and CLV2-SOL2/CRN perceive the CLV3 peptide ligand. The suppression of WUS expression leads to the restriction of the stem cell population in the SAM and balances cell proliferation and differentiation. Recently we isolated RPK2 as a third receptor of the CLV3 signal in Arabidopsis. So, at least three receptors, CLV1, CLV2-SOL2/CRN and RPK2 are responsible for CLV3 perception for SAM maintenance system in Arabidopsis.Citation3–Citation7
CLE peptides are considered to function in plant morphogenesis as intercellular signaling molecules,Citation8–Citation14 and CLE genes have been found in various seed plants and even in the moss Physcomitrella patens.Citation2,Citation15–Citation18 As for the receptors, CLV1 homologues were identified not only in vascular plants but also in the bryophyte Physcomitrella.Citation18 However, CLV2 and SOL2/CRN homologues were found only in higher plants, and were not found in the genomes of Selaginella moelendorffii or Physcomitrella patens.Citation18
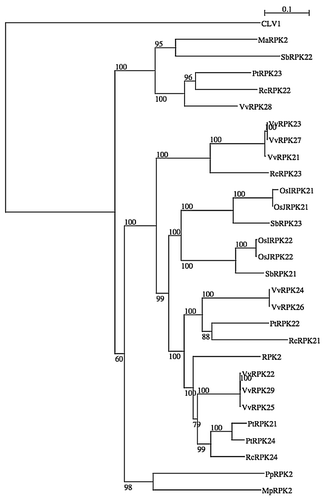
To identify RPK2 homologues in plants, we performed BLAST search. RPK2 homologues were found not only from seed plants but also from moss, Physcomitrella patens, and from liverwort, Marchantia polymorpha. Only one homolog was found from moss and liverwort. However, most of other higher plants have multiple homologues. This result indicates that the RPK2 homologues are responsible for many physiological events in higher plants.
Our phylogenetic analysis indicates that RPK2 and CLV1 have evolved from early evolutional steps in plants. On the other hand, CLV2 and SOL2/CRN homologues were found as a single gene only in higher plants, indicating that they have specific functions in multicellular organisms. CLV2 and SOL2/CRN might be necessary to evolve complex multicellular SAM structure in plants.
Functional diversity of RPK2 and CLV1 homologues remains to be resolved. The existence of a number of RPK2 and CLV1 homologues among higher plants implies a functional diversity among them. Except for the functions of CLV3 and TDIF, the functions of most CLE peptides are largely unknown. The direct interaction between CLE peptides and their cognate receptors will be examined with a biochemical approach because it is possible that these receptors bind other ligands. Further genetic and biochemical studies of these receptor-like proteins in various plant species will provide further evidence for CLE peptide and receptor functions in various aspects of plant development.
Abbreviations
| CLE | = | CLAVATA3/ESR-related |
| SAM | = | shoot apical meristem |
| RAM | = | root apical meristem |
| WUS | = | wuschel |
| CLV | = | clavata |
| LRR | = | leucin rich repeat |
| CRN | = | coryne |
| SOL2 | = | suppressor of LLP1 2 |
| RPK2 | = | receptor like protein kinase 2 |
| MaRPK2 | = | Musa acuminata RPK2-like protein |
| OsIRPK2 | = | Oryza sativa indica group RPK2-like protein |
| OsJRPK2 | = | Oryza sativa japonica group RPK2-like protein |
| PtRPK2 | = | Populus trichocarpa RPK2-like protein |
| VvRPK21 | = | Vitis vinifera RPK2-like protein |
| SbRPK2 | = | Sorghum bicolor RPK2-like protein |
| RcRPK2 | = | Ricinus communis RPK2-like protein |
| PpRPK2 | = | Physcomitrella patens RPK2-like protein |
| MpRPK2 | = | Marchantia polymorpha RPK2-like protein |
Figures and Tables
Figure 1 A neighbor-joining tree of RPK2. Protein sequence of RPK2 homologues of Arabidopsis thaliana (RPK2 and CLV1 as an out group), Musa acuminata (MaRPK2: ABF72006.1), Oryza sativa Indica Group (OsIRPK21: EAY91907.1, OsIRPK22: EAZ04623.1), Oryza sativa Japonica Group (OsJRPK21: NP_001051315.1, OsJRPK22: NP_001060207.1), Populus trichocarpa (PtRPK21: XP_002305358.1, PtRPK22: XP_002311344.1, PtRPK23: XP_002321080.1, PtRPK24: XP_002323902.1), Vitis vinifera (VvRPK21: CAN67126.1, VvRPK22: CAN77668.1, VvRPK23: CAO40245.1, VvRPK24: CAO41857.1, VvRPK25: CAO49170.1, VvRPK26: XP_002274047.1, VvRPK27: XP_002274211.1, VvRPK28: XP_002276030.1, VvRPK29: XP_002279979.1), Sorghum bicolor (SbRPK21: XP_002460974.1, SbRPK22: XP_002462751.1, SbRPK23 XP_002463860.1), Ricinus communis (RcRPK21: XP_002512071.1, RcRPK22: XP_002512822.1, RcRPK23: XP_002515143.1, RcRPK24: XP_002527617.1), Physcomitrella patens (PpRPK2: XP_001781758.1) and Marchantia polymorpha (MpRPK2: isotig22424), were used to construct the phylogenic tree. Bootstrap values of 60% and above, from the neighbor-joining method with Kimura's correction, are shown. The scale bar indicates the number of amino acid substitutions per site.
Addendum to:
References
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 2006; 313:845 - 848
- Kinoshita A, Nakamura Y, Sasaki E, Kyozuka J, Fukuda H, Sawa S. Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol 2007; 48:1821 - 1825
- Miwa H, Kinoshita A, Fukuda H, Sawa S. Plant meristems: CLAVATA3/ESR-related signaling in the shoot apical meristem and the root apical meristem. J Plant Res 2009; 122:31 - 39
- Miwa H, Betsuyaku S, Iwamoto K, Kinoshita A, Fukuda H, Sawa S. The receptor-like kinase SOL2 mediates CLE signaling in Arabidopsis. Plant Cell Physiol 2008; 49:1752 - 1757
- Müller R, Bleckmann A, Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 2008; 20:934 - 946
- Kinoshita A, Betsuyaku S, Osakabe Y, Mizuno S, Nagawa S, Stahl Y, et al. RPK2/TOAD2 is an essential receptor-like kinase transmitting the CLV3 signalling in Arabidopsis. Development 2010; 137:3911 - 3920
- Betsuyaku S, Takahashi F, Kinoshita A, Miwa H, Shinozaki K, Fukuda H, Sawa S. Mitogen-Activated Protein Kinase regulated by the CLAVATA receptors contributes to the shoot apical meristem homeostasis. Plant Cell Physiol 2010; 52:14 - 29
- Chu H, Qian Q, Liang W, Yin C, Tan H, Yao X, et al. The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol 2006; 142:1039 - 1052
- Fiers M, Golemiec E, van der Schors R, van der Geest L, Li KW, Stiekema WJ, Liu CM. The CLAVATA3/ESR motif of CLAVATA3 is functionally independent from the nonconserved flanking sequences. Plant Physiol 2006; 141:1284 - 1292
- Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 2006; 313:842 - 845
- Ni J, Clark SE. Evidence for functional conservation, sufficiency and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol 2006; 140:726 - 733
- Sawa S, Kinoshita A, Nakanomyo I, Fukuda H. CLV3/ESR-related (CLE) peptides as intercellular signaling molecules in plants. Chem Rec 2006; 6:303 - 310
- Strabala TJ, O'donnell PJ, Smit AM, Ampomah-Dwamena C, Martin EJ, Netzler N, et al. Gainof-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol 2006; 140:1331 - 1344
- Suzaki T, Toriba T, Fujimoto M, Tsutsumi N, Kitano H, Hirano HY. Conservation and diversification of meristem maintenance mechanism in Oryza sativa: Function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol 2006; 47:1591 - 1602
- Sharma VK, Ramirez J, Fletcher JC. The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Mol Biol 2003; 51:415 - 425
- Oelkers K, Goffard N, Weiller GF, Gresshoff PM, Mathesius U, Frickey T. Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol 2008; 8:1
- Sawa S, Kinoshita A, Betsuyaku S, Fukuda H. A large family of genes that share homology with CLE domain in Arabidopsis and rice. Plant Signal Behav 2008; 3:337 - 339
- Miwa H, Tamaki T, Fukuda H, Sawa S. Evolution of CLE signaling. Plant Signal Behav 2009; 4:477 - 481