Abstract
Plants are capable of perceiving changes in the light environment and finely adjust their growth and development. Reductions of red to far-red ratio (R:FR) generated by an increase of the plant canopy above the plant are sensed by the phytochrome system triggering the shade-avoidance syndrome (SAS) that includes elongation of vegetative structures, reduction of branching and acceleration of flowering. Albeit the SAS is a strategy of major adaptative significance in plant communities, involving massive changes in gene expression, our knowledge of the SAS signaling network is still fragmented. By a selection and characterization of a T-DNA mutant with a long hypocotyl under shade, we identified BBX21, a protein with two B-box domains involved in the SAS. BBX21 belongs to a small eight member family of B-box containing proteins with both opposite and additive functions in the SAS signaling. BBX21 down-regulates the gene expression of auxin, brassinosteroid and ethylene signaling pathway components under shade. Furthermore BBX21 is a transcription factor that interacts genetically with COP1. We propose a model in which a dynamic balance of positive and negative B-box transcriptional regulators acts as a gas-and-brake mechanism into the COP1 signaling to regulate the expression of SAS.
Plants monitor the spectral quality, fluence, direction and duration of light by different photosensory systems. Phytochromes sense red light (R; 580–690 nm) and far-red light (FR; 690–800 nm); and cryptochromes, phototropins and zeitlupes perceive blue light (B; 380–495 nm) and UV-A (320–380 nm). In shaded environments, the light is relatively enriched in FR due to the selective absorption of B and R by chlorophyll pigments that reduces the R:FR ratio, a signal that is finely perceived and transduced by the phytochromes. Low R:FR reduces the proportion of Pfr, the biologically active FR-absorbing form of the phytochromes, in plant tissues and consequently triggers different responses known as the shade-avoidance syndrome (SAS) that includes elongation of hypocotyls, petioles and stems, reduction of branching and acceleration of flowering.Citation1
The reduction of the R/FR ratio activates a complex signaling network.Citation1 A group of transcription factors are rapidly upregulated upon exposure of plants to low R:FR such as ATHB2, a homeodomain-leucine zipper (HD-Zip) factor; and PIF4 and PIF5, two bHLH like proteins, that increase their stability at low R:FR and act as positive regulators of SAS.Citation2,Citation3 In contrast, other bHLH transcription factors such as HFR1, PAR1 and PAR2, are early negative modulators of SAS preventing an exaggerated expression of SAS.Citation4,Citation5 Furthermore, the DELLA family, a set of giberellin signaling repressing proteins, are degraded under low R:FR allowing the elongation of plant structures.Citation6,Citation7 Sixteen years ago, McNellis showed that cop1 mutant seedlings display null SAS phenotype suggesting a positive function of COP1 in shade conditions.Citation8 More recently, Roig-Villanova demonstrated that COP1 modulates SAS gene expression in response to simulated shade in light-grown seedlings.Citation9 In addition to bHLH and HD-Zip transcription factors, B-box-containing proteins (BBX) also act as regulators of light signaling in plants, and some of them interact into the COP1 transcriptional complex.Citation10–Citation12 All these experimental evidences suggest a potential functional role of BBX under shade. Here we demonstrated an undocumented function of BBX in the regulation of plant growth under canopy shade and low R:FR ratio through the COP1 signaling.
Complex and Antagonist Roles of BBX for the Regulation of Plant Growth Under Shade
The B-box-containing proteins are a family of 32 members classified in five structure groups.Citation13 The group IV of BBX proteins, containing two B-boxes, B1 and B2 but lacking a CCT domain, is represented by eight members (BBX18 to BBX25). By phenotypic and gene expression analysis of bbx mutants of the group IV in the SAS, we conclude that (1) different BBX members show antagonist functions (BBX19, BBX21 and BBX22 are repressors, and BBX18 and BBX24 are promoters); (2) some of them have prominent roles in the regulation of plant growth under shade (BBX21 and BBX24); (3) some members of BBX show additive effects (BBX21 and BBX22); and (4) gene expression of BBX that belongs to other structure groups are also differentially regulated by shade ().
BBX21 Downregulates the Gene Expression of Auxins, Brassinosteroids and Ethylene Under Shade
To understand the molecular basis of the physiological alterations of bbx21 mutant in SAS, we used Affymetrix ATH1 microarrays to compare the transcriptome of wild-type and bbx21 seedlings exposed to sunlight or canopy.Citation14 Global expression analysis identified a group of genes regulated by BBX21 specifically under canopy shade. The analysis found 576 genes whose expression was statistically different between wild-type and bbx21 being 205 genes downregulated by BBX21. By comparison with a transcriptional analysis of hormonal regulation in Arabidopsis seedlings,Citation15 we found a significant association between genes downregulated by BBX21 under shade and those upregulated by auxins, brassinosteroids and ethylene (). These results suggest that BBX21 acts as a negative regulator of SAS in conjunction with specific hormonal transcriptional networks.
BBX21 and BBX22 Mediate Shade Responses through COP1 Signaling
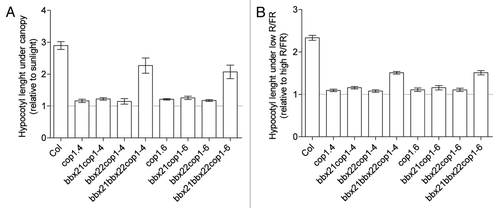
We evaluated the function of BBX21 and BBX22 in the COP1 signaling network regulating SAS by the phenotype analysis of bbx21, bbx22 and cop1 single, double and triple mutants. While both bbx21cop1 and bbx22cop1 resembled the phenotype of cop1–4 and cop1–6, the bbx21bx22cop1 triple mutants restored, at least partially, the wild-type phenotype under canopy and low R/FR, the most prominent light signal in shaded conditions (). Expression analysis of early SAS genes also suggests that BBX21 and BBX22 are components involved in the COP1 signaling probably as part of a common protein complex.
Conclusions
The B-box-containing proteins constitute a diverse and conserved group of transcription factors present in plants and other eukaryote organisms. Here we show that BBX21 is a negative regulator of the SAS acting, at least partially, downregulating the expression of genes associated with auxin, brassinosteroid and ethylene signaling pathways. Furthermore BBX21 and BBX22 interact genetically with COP1 probably into a molecular complex that regulates SAS at the transcriptional level. The B-box domains of BBX21 and BBX22 are though to facilitate the protein-protein interactions such as those occurring in transcriptional complexes.Citation13 Besides other members of BBX play opposite roles to BBX21 and BBX22 suggesting that different BBX may be involved into the COP1 signaling to regulate the expression of SAS through a gas-and-brake mechanism that prevents exaggerated expression of SAS.
Abbreviations
| R:FR | = | red to far-red ratio |
| R | = | red light |
| B | = | blue light |
| FR | = | far-red light |
| SAS | = | shade-avoidance syndrome |
| Pfr | = | FR-absorbing form of the phytochromes |
| BBX | = | B-box containing protein |
| COP1 | = | constitutive photomorphogenic1 |
Figures and Tables
Figure 1 Complex and antagonist roles of BBX for the regulation of plant growth under shade. (A) Hypocotyl length of wild-type and bbx mutants of group IV seedlings grown under canopy with respect to sunlight. (B) qRT-PCR analysis of BBX genes of group IV in the wild-type in response to canopy light. The expression of transcripts was normalized to the IPP2 gene, and data was standardized to sunlight. (C) Microarrays expression data of other structure groups of BBX genes in wild-type in response to canopy light. The expression of each gene under canopy was standardized to sunlight.
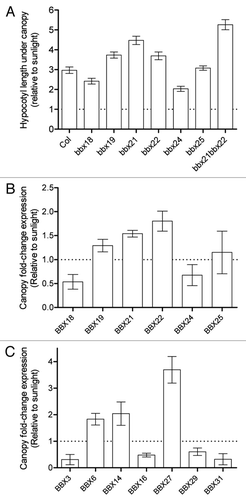
Figure 2 BBX21 downregulates the gene expression of auxins, brassinosteroids and ethylene under shade. Venn diagram shows genes upregulated in bbx21 seedlings under canopy that are significant associated with hormone genes expressed in the auxin, ethylene and brassinosteroid signaling networks during seedling development.
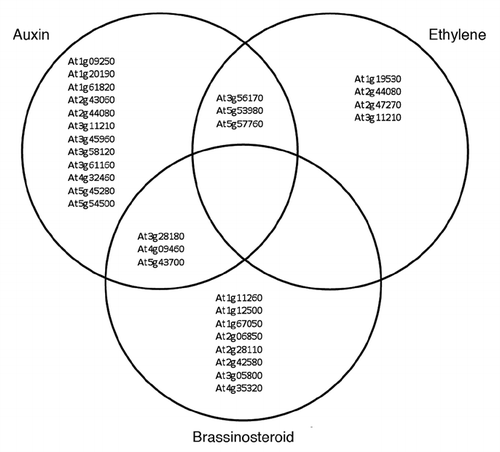
Figure 3 BBX21 together with BBX22 restore partially the SAS phenotype in the cop1 background. (A) Hypocotyl length of wild-type, cop1, bbx21cop1, bbx22cop1 and bbx21bbx22cop1 seedlings grown under canopy (relative to sunlight) and (B) under low R/FR = 0.35 (relative to high R/FR = 3.4) in a growth light chamber.
Acknowledgements
We thank the University of Buenos Aires (Grant G041 to J.F.B.) and the Agencia Nacional de Promoción Científica y Tecnológica (PICT 2008-1061 to J.F.B.).
Addendum to:
References
- Franklin KA. Shade avoidance. New Phytol 2008; 179:930 - 944
- Carabelli M, Morelli G, Whitelam G, Ruberti I. Twilight-zone and canopy shade induction of the Athb-2 homeobox gene in green plants. Proc Natl Acad Sci USA 1996; 93:3530 - 3535
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 2008; 53:312 - 323
- Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, et al. A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev 2005; 19:2811 - 2815
- Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L, Portolés S, Rodríguez-Concepción M, Martínez-García JF. Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J 2007; 26:4756 - 4767
- Achard P, Gusti A, Cheminant S, Alioua M, Dhondt S, Coppens F, et al. Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol 2009; 19:1188 - 1193
- Djakovic-Petrovic T, de Wit M, Voesenek LA, Pierik R. DELLA protein function in growth responses to canopy signals. Plant J 2007; 51:117 - 126
- McNellis TW, von Arnim AG, Deng XW. Overexpression of Arabidopsis COP1 results in partial suppression of light-mediated development: evidence for a light-inactivable repressor of photomorphogenesis. Plant Cell 1994; 6:1391 - 1400
- Roig-Villanova I, Bou J, Sorin C, Devlin PF, Martinez-Garcia JF. Identification of primary target genes of phytochrome signaling. Early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol 2006; 141:85 - 96
- Kumagai T, Ito S, Nakamichi N, Niwa Y, Murakami M, Yamashino T, Mizuno T. The common function of a novel subfamily of B-box zinc finger proteins with reference to circadian-associated events in Arabidopsis thaliana. Biosci Biotechnol Biochem 2008; 72:1539 - 1549
- Datta S, Hettiarachchi C, Johansson H, Holm M. SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 2007; 19:3242 - 3255
- Datta S, Johansson H, Hettiarachchi C, Irigoyen ML, Desai M, Rubio V, Holm M. LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 2008; 20:2324 - 2338
- Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, Wu SH. The Arabidopsis B-box zinc finger family. Plant Cell 2009; 21:3416 - 3420
- Crocco CD, Holm M, Yanovsky MJ, Botto JF. AtBBX21 andCOP1 genetically interact in the regulation of shade avoidance. Plant Journal 2010; 64:551 - 565
- Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 2006; 126:467 - 475