Abstract
Plants have evolved adaptations to environmental factors, including UV-B present in solar radiation. Deployment of specific adaptive phenotypes to avoid or repair UV-B damage requires physiological and developmental acclimation to variable UV-B fluence. To gain a better understanding of the events in UV-B acclimation, we have analyzed a 5min to 6h time course of transcriptome and metabolome responses in irradiated and shielded leaves and in immature maize ears to unravel the systemic physiological and developmental responses in exposed and shielded organs. Within 10 min of UV-B exposure, transcripts are changed not only in irradiated leaves, but also in shielded tissues. The number of UV-B-regulated transcripts rapidly increases with exposure length. Interestingly, after 10 min of exposure, the overlap in transcriptome changes in irradiated and shielded organs is significant; while, after 6h of UV-B, most transcripts are specific for each tissue under study. We suggest that early events in all tissues may be elicited by common signaling pathways, while at longer exposure times responses become more organ-specific. Our working hypothesis is that mobile signaling molecules are generated in irradiated organs to elicit the initial responses. We found several metabolites that rapidly change after different treatments during the timecourse; myoinositol is one candidate metabolite based on its rapid modulation in all organs. There is also support from RNA profiling: after 1h UV-B, transcripts for myoinositol-1-phosphate synthase are decreased in both irradiated and shielded leaves suggesting downregulation of biogenesis.
UV-B Effects in Plants
Solar radiation contains light qualities that are essential for photosynthesis and development, but certain wavelengths can also damage cells. Plants contain multiple photoreceptors: phytochromes for perceiving red/far red, cryptochromes and phototropins for blue/UV-A, and at least one UV-B receptor, UVR8, which was recently identified.Citation1 The physiological mechanisms that coordinate local and systemic responses to UV-B photons remain largely unknown for both low UV-B exposure and high fluence that can cause cellular damage by generating photoproducts in DNA and by direct damage to proteins, lipids, and RNA.
Under normal solar fluence, there is a balance of damage and the repair or replacement of these macromolecules. Maize lines show differential capacity to avoid damage from ambient UV-B, primarily reflecting differences in flavonoid sunscreen content: higher levels of flavonoids are correlated with fewer stress responses to UV-B.Citation2 Indeed, landraces from high altitude sites exhibit greater tolerance to UV-B, and this property is correlated with both higher flavonoidsCitation3 and greater chromatin remodeling capacity;Citation4 conversely, lines with knockouts in factors associated with chromatin remodeling are hypersensitive to UV-B in adult tissue and exhibit seedling lethality.Citation4 In addition to ambient UV-B, the depletion of the ozone layer results in periodic, but unpredictable spikes in UV-B intensity in the polar and temperate zones. Consequently, determining the molecular basis for UV-B acclimation to normal fluence rates and the capacity to tolerate UV-B fluence spikes is an important factor in sustaining crop yield. Thus, our focus is to discover the signals produced in UV-B-irradiated leaves that elicit physiological and developmental changes in shielded organs such as immature leaves and ears. Systemic responses have the potential to impact yield by modulating ear growth or kernel properties, in addition to the “short-term” and readily repaired damage to DNA and photosynthetic reaction centers in irradiated leaves. Therefore, tracking response kinetics to elevated UV-B irradiation in sunlit leaves and identifying the signals produced there that subsequently elicit systemic changes in reproductive organs should elucidate how UV-B affects plant yield.
Previously, we found that shielded organs such as immature ears exhibit significant transcriptome changes within two hours of UV-B irradiation of leaves, with the first detectable changes at 1 h.Citation5 Furthermore, a key result of prior studies is that leaves exposed to UV-B ~3-fold higher than ambient solar noon for brief periods (1–2 h) generate signals that result in transcriptome changes in completely shielded distant organs such as immature ears and leaves wrapped in UV-B filters.Citation5 Now our goal is to unravel the systemic physiological and developmental responses maize exhibits to excess UV-B in both irradiated and shielded organs. To accomplish this, we conducted two parallel lines of investigation: transcriptome profiling to quantify the kinetics of alterations in exposed and shielded organs and metabolic profiling in irradiated tissues to elucidate possible signaling molecules and their appearance in shielded organs.
Transcriptome Analysis During a Time-Course
To gain insight into the kinetics of transcriptome responses in UV-B-irradiated maize plants, the uppermost two leaves of 5–6 week-old greenhouse grown plants were irradiated for 10 min, 30 min, 1, 2 or 6 h for an analysis of the dynamics of responses.Citation6,Citation7 For this purpose, adult maize plants were covered with a plastic sheath that absorbs UV-B (polyester, PE), while two adult leaves per plant were irradiated with UV-B radiation through a plastic that allows UV-B transmittance (CA, for details, seeCitation6). The transcriptomes from irradiated and shielded leaves (IL and SL, respectively), and immature ears (IE) were analyzed using microarrays to identify early and late UV-B responses. shows that after 10 min of UV-B irradiation, 262 transcripts are changed by more than 2-fold (p < 0.05) in IL, 148 in SL and 73 in IE. After 1h of UV-B, the number of transcript changes increases by 2-fold in IL, 2.35-fold in SL, and 8.37-fold in IE; while after 6h,the number of mRNAs regulated by UV-B is5.1-times higher than after 10 min in IL, 8.65 in SL and 6.29 in IE. Therefore, although it is clear that transcriptome changes occur as early as after 10 min of irradiation, more responses occur with longer exposure. Moreover, transcript categories that are UV-B-regulated at early time points differ from those changed at longer times of exposure as previously reported.Citation6,Citation7 For example, transcription factors and signal transduction proteins are highly represented in our transcriptome analysis after 10 min irradiation,Citation7 while transcripts that encode genes in secondary metabolism like phenylalanine ammonia-lyase, flavonoid 3–hydroxylase, cinnamoyl-CoA reductase, which all participate in the phenylpropanoid pathway for the synthesis of flavonoids and cell wall, are all induced by UV-B at longer times of exposure;Citation6 these are not significantly represented in our classification for 10, 30, and 60 min exposure times.Citation7
Figure 1. Venn diagrams comparing transcriptome changes in leaves and immature ears from plants irradiated with two canopy leaves exposed compared with non-irradiated control plants. Only two adult leaves per plant were irradiated over a time course of 10, 30 min, and 1, 2, 4, and 6 h. (A) Intersection of genes differentially expressed in irradiated leaves (IL); (B) Intersection of genes differentially expressed in shielded leaves (SL); (C) Intersection of genes differentially expressed in immature ears (IE). Each sample was compared with plants under control conditions in the absence of UV-B (C). Transcripts showing changes higher than 2-fold (p < 0.05) were included in the classification.
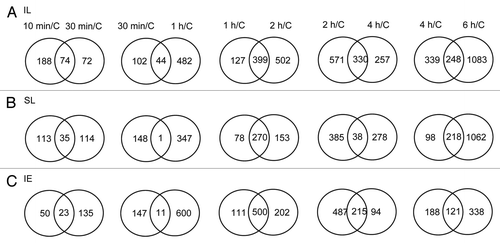
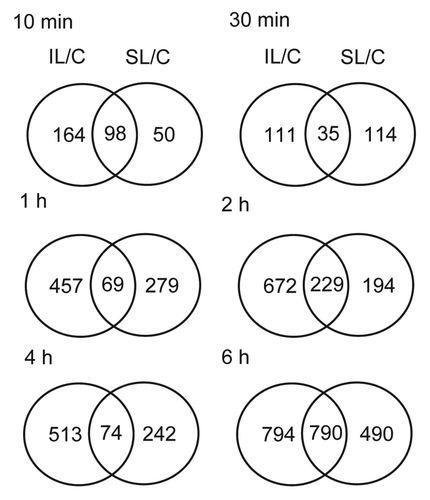
On the other hand, there are 1.77-fold more transcripts changed by UV-B in IL than in SL after 10 min of treatment (), this difference slightly decreases after 1h (1.51 times higher) and declines further after 6h (1.04 times higher); this suggests that at very early times of exposure, irradiated tissues respond faster than shielded ones, while after longer times, all tissues show a similar magnitude of transcriptome changes. The specificity of organ responses increases with exposure length over the first hour (). For example, after 10 min of UV-B, there are 98 transcripts that are UV-B-regulated similarly between IL and SL. The percentage of overlapping transcripts decreases with time of exposure, after 30 min there is only 13.4% of overlap between transcriptome changes, while after 1h 8.5% of the UV-B-regulated transcripts in IL are similarly regulated in SL (). This result suggests that, initially, there are common transcriptome responses; but over the subsequent hour organs show more distinctive responses. Ultimately, many of the transcripts altered in abundance by UV-B are common. This is clear after 6h of UV-B, where 50% of the transcripts changed in IL are similarly changed in SL (). As previously reported,Citation6 many transcripts that show changes at this late time point correspond to genes in stress responses, primary and secondary metabolism and photosynthesis; these changes correspond to general stress responses in irradiated and shielded tissues, such as leaves and immature ears.
Figure 2. Venn diagrams comparing transcriptome changes in IL and SL from UV-B-irradiated plants compared with non-irradiated control plants (C). Plants were irradiated over a time course of 10, 30 min, and 1, 2, 4 and 6h. Intersection of genes differentially expressed after the different time points in IL and SL. Transcripts showing changes higher than 2-fold (p < 0.05) were included in the classification.
shows that there are 73 transcripts that are changed by at least 2-fold (p < 0.05) in immature ears after 10 min of UV-B irradiation; this number more than doubles after 30 min of UV-B exposure. Of the 73 transcripts that are changed after 10 min, 32% also show altered expression after 30 min of UV-B irradiation (central element of the Venn diagram, ), while 50 transcripts are only changed after 10 min UV-B; these may be involved in early responses to UV-B in this organ and probably correspond to proteins that participate in early UV-B signaling. After longer UV-B exposure times, similarly as found in leaves, the number of regulated transcripts increases. An interesting observation is that at time points longer than 1h, there is upregulation of transcripts that encode for enzymes in cell wall degradation: endo-1,4-β-glucanases, β-fructofuranosidases, β-glucosidases, β-1,3-glucanases, and polygalacturonases.Citation6 The upregulation of these genes is specific for immature ears, so it is possible in these organs UV-B induces some specific cell wall reorganization. One possibility is that in this organ, UV-B radiation may be affecting organ growth, a plausible mechanism for impacting yield.
It is interesting that some of the UV-B-regulated transcripts at 10 min are not significantly altered by UV-B at longer irradiation times.Citation7 These probably correspond to genes that encode proteins that participate in early responses to UV-B in maize: in this group, there are 29 DNA binding proteins including transcription factors and 7 proteins that participate in signal transduction.Citation7 In the list of overlapping transcripts between IL and SL at this time point, 19 correspond to transcription factors/DNA binding proteins; therefore, the signal that induces transcription must be transmitted quickly to the shielded leaves. In immature ears, 3 transcription factors are also UV-B-regulated after 10 min of UV-B (heat shock factor RHSF6, an ethylene-responsive transcriptional coactivator-like protein, and an ethylene-responsive transcription factor 3) and are shared with irradiated and shielded leaves. In addition, there are 24 transcripts common among the 3 organs after 10 min UV-B.Citation7 It is interesting to note that 7 correspond to heat shock proteins. After longer UV-B exposure times, a number of HSPs are downregulated by UV-B in IE.Citation6 Together, our results suggest that changes in the expression levels of this group of proteins may have an important role in UV-B responses, both in irradiated and shielded tissues, and transient changes in their levels would be important for acclimation to this radiation.
In addition, at short times of UV-B exposure (less than an hour), the classification of UV-B-regulated transcripts by GO categories show that the transcription/transcriptional regulation category has the highest representation.Citation7 In this category, the number of mRNAs in this group is significantly increased after 1h UV-B in all tissues, prerequisite to the major reprogramming of gene expression that occurs over the following hours in continuous UV-B.Citation6 On the other hand, for the signal transduction category, the number of transcripts changed after 10 min irradiation is higher in leaves than in ears, with more transcripts changed in IL than in SL. Thus, UV-B-signaling is more prominent in tissues that are directly exposed to UV-B than in shielded ones. As for transcripts in secondary metabolism, a similar differential expression at shorter and longer UV-B exposure times is observed for other categories, such as DNA metabolism including DNA repair. Genes involved in DNA repair metabolism are induced at longer times of exposure, when damaged DNA is accumulated, and this mostly occurs in organs that are directly irradiated with UV-B.Citation6
How is UVR8, a proposed UV-B photoreceptor in Arabidopsis,Citation1 regulated in maize? This transcript is initially increased in irradiated and shielded tissues.7At longer UV-B exposure times (2h), maize UVR8 is downregulated both in IL and SL.Citation6 UVR8 is a UV-B-specific signaling component that mediates low fluence photomorphogenic responses, and it is required for UV-B induced expression of the gene encoding the HY5 transcription factorCitation8,Citation9 in co-operation with COP1. UVR8 and COP1 interact directly and rapidly in the nucleus after UV-B exposure;Citation10 this is a very early step in UV-B signaling responses, ensuring UV-B acclimation and protection.Citation11 In Arabidopsis, UV-B signaling by UVR8 is repressed by two highly related WD40-repeat proteins, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 and 2 (RUP1 and RUP2); both genes are transcriptionally activated by UV-B in a COP1-, UVR8-, and HY5-dependent manner.Citation12 RUP1 and RUP2 act downstream of UVR8–COP1 in a negative feedback loop impinging on UVR8 function, balancing UV-B defense measures with plant growth. However, no homologs of either RUP1 or RUP2 are present in the maize genome. Thus, a possibly different UV-B signaling pathway may be present in maize. Despite this, if UVR8 is a UV-B sensor in maize, turning down responses appears to be important for successful acclimation.
Identification of UV-B-Induced Metabolomic Changes
As a first step in identifying potential signal molecules moving from irradiated leaves to shielded organs, we conducted metabolic profiling using GC-MS to find metabolites altered by UV-B radiation over a time course in IL and SL. Because transcriptome analysis identified changes within 10 min, metabolite samples were analyzed after 5, 10, 15, 30, 60, 90 min and 2, 4 and 6h of UV-B irradiation for comparison to untreated control plants (no UV-B). We identified 84 compounds, 22 of which had a statistically significant change in our two leaf irradiation protocol (, one way ANOVA). Six metabolites were only changed in irradiated leaves (leucine, fructose, glucose, shikimic acid, quinic acid, and trans-caffeoylquinic acid); these include three compounds in the phenylpropanoid pathway (shikimic acid, quinic acid, and trans-caffeoylquinic acid; ). In addition, UV-B-regulated genes in the flavonoid pathway show increased levels exclusively in directly exposed leaves,Citation6 suggesting that these metabolites are not translocated to shielded tissues nor do mobile signals induce them in shielded organs.
Figure 3. Metabolic profiling from UV-B-irradiated leaves. Metabolites from two irradiated leaves (IL) and SL covered with a plastic sheath that absorbs UV-B were analyzed after 5, 10, 15, 30, 60, 90 min and 2, 4 and 6 h exposure. As a control, samples from non-irradiated leaves (no UV-B) were included. Statistical analysis was done using one way ANOVA; statistically significant differences are labeled with letters a, b, c and d (α = 0.05).
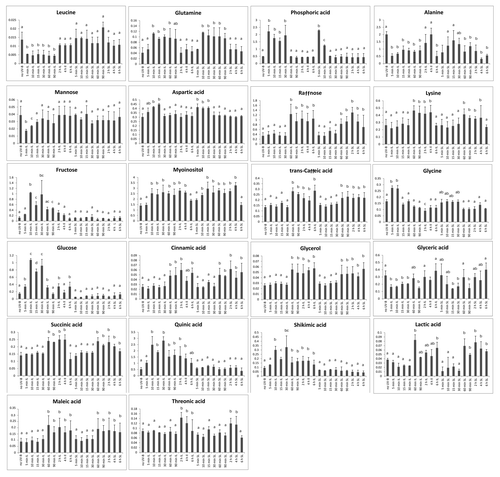
Because a signaling metabolite(s) must increase quickly in irradiated leaves to trigger transcriptome changes in shielded organs within an hour, we predicted that such molecules would show 1) high concentrations in treated leaves relative to untreated plants and 2) increases in shielded organs. Of the 22 metabolites changed by UV-B (), 13 of these had a statistically significant change by UV-B at exposure times less than 1.5 h (, one way ANOVA). Five metabolites were increased in both IL and SL (aspartic, phosphoric, and glyceric acids, glutamine, and myoinositol, ) while changes in eight metabolites were restricted to IL: alanine, fructose, glucose, glycine, leucine, mannose, shikimic acid, and quinic acid (). Metabolites modulated by UV-B in both IL and SL are potential signal molecules synthesized in exposed leaves and translocated to shielded organs; or alternatively, an unknown signal could be transmitted to shielded tissues, and this signal could induce the synthesis of these compounds in shielded tissues. Myoinositol is of particular interest in light of our microarray results.Citation6 We reported that transcripts for myoinositol-1-phosphate synthase were downregulated by UV-B in both IL and SL after 4h of UV-B irradiation,Citation6 which would be predicted to increase the levels of the precursor myoinositol. Either lowered levels of myoinositol-1-phosphate or elevated myoinositol could be signaling molecules coordinating UV-B responses. shows that myoinositol levels are rapidly increased after 10 min of UV-B both in IL and SL, confirming it as a UV-B-signaling candidate. Myoinositol is synthesized from glucose-6-P, which is converted to myoinositol-1-phosphate by myoinositol-1-phosphate synthase, and this compound is dephosphorylated to produce myoinositol. The step catalyzed by the synthase is rate-limiting for myoinositol biosynthesis in plants.Citation13,Citation14 Myoinositol is a candidate UV-B signaling compound, because it is rapidly increased in irradiated and shielded leaves. Rapid synthesis after brief exposure is expected for a signal, while longer irradiation (4h or longer) could provoke a downregulation to modulate metabolite levels. Myoinositol has been proposed as important in stress protection,Citation15 and may also regulate programmed cell death in Arabidopsis. Mutants in one myoinositol-1-phosphate synthase (AtIPS1) exhibit accelerated cell death; the encoded protein has a nuclear localization sequence, suggesting that nuclear pools of myoinositol may be critical.Citation16
It is interesting that 9 of the 22 UV-B-modulated metabolites are increased at short time points and return to their normal level in the absence of UV-B (leucine, glutamine, phosphoric acid, fructose, glucose, glycine, glyceric acid, aspartic acid and mannose, ), while 4 metabolites are changed at early exposure times and remain changed during longer exposure times (alanine, quinic acid, myoinositol and shikimic acid, ). These latter changes reflect global metabolic changes that are induced by UV-B.
Many of the measured metabolite changes are accompanied with changes in transcript levels of their metabolizing enzymes. For example, the increase in Gln levels at short UV-B exposure times () parallel an increase in the levels of mRNAs for Gln synthetase, both the cytosolic (GRMZM2G036464) and the chloroplastic (GRMZM2G098290) isoforms at 10 min of UV-B exposure (see GEO under ID GSE25038). At longer exposure times (1h), both transcripts show a decrease by UV-B compared with levels in untreated plants, and this decrease is accompanied by a decrease in Gln at exposure times longer than 90 min (). Similarly, while Asp levels are increased at very short UV-B exposure times and return to basal levels at longer exposure times (), transcripts for an Asp aminotransferase (GRMZM2G033799) are decreased after 1 h of UV-B treatment (see GEO under ID GSE25038). In addition, sugars like fructose and glucose also show rapid increases in their levels by UV-B and return to basal levels at exposure times longer than 90 min and 60 min, respectively (). The phosphorylated forms of these sugars were not detected in our metabolome experiments; it is possible that they are hydrolyzed by the protocol used. If this is true, the changes in the expression levels of some transcripts in glycolysis also show similar patterns as those observed for the amino acids Gln and Asp. For example, transcripts for phosphoglucomutase (GRMZM2G025854), and fructose 1,6 biphosphate aldolase (GRMZM2G057823) are decreased after 1h UV-B in parallel to the decrease measured for the 2 sugars (). Sugar translocators such as a glucose (GRMZM2G004694) and a glucose-6-P translocator (GRMZM2G125850) show increased levels after 1h of UV-B, suggesting that changes in this sugar localization may be important in UV-B signaling.
In summary, our protocol has generated a list of metabolites changed by UV-B in irradiated leaves and that are either translocated to shielded leaves or elevated there by an unknown signal. One or more of these may act as signaling compounds in shielded organ responses to UV-B in maize. Defining the signaling molecules will be a first step in understanding how systemic responses are triggered and coordinated.
Conclusions
Our focus was to document the scope and kinetics of systemic changes in shielded leaves and immature ears after irradiation of two canopy leaves in adult plants and to compare irradiated and shielded organs over a time course. Transcriptome profiling was used to track macromolecular alterations in exposed and shielded organs. In parallel, metabolome profiling was used to search for candidate signaling molecules. Our experiments have demonstrated that direct exposure of just the top leaves substantially alters the transcriptome of both irradiated and shielded organs. As little as 10 min of UV-B causes substantial transcriptomic changes not only in irradiated but also in shielded tissues. Moreover, after 5 min, 9 metabolites change quickly in irradiated leaves relative to untreated plants and also increased in shielded organs (leucine, phosphoric acid, alanine, quinic acid, fructose, glucose, glycine, glyceric acid, and mannose, ). In this study, candidates for components of signal transduction and possible signal molecules were identified; for example, myoinositol or a derivative may be (a) signaling candidate(s) in UV-B responses, as this compound is increased by UV-B after 10 min in irradiated and shielded leaves (). Collectively, the results presented here highlight possible signaling pathways and molecules for future research. An important next step will be understanding of regulatory networks that permit acclimation responses to UV-B.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Mónica Hourcade for GC/MS technical support. Research supported by USDA National Research Initiative Grant 2008–35100–04578 to V.W. and by FONCyT grants PICT-2006–00957 and PICT-2007–00711 to P.C. P.C. is a member of the Researcher Career of CONICET.
References
- Rizzini L, Favory J-J, Cloix C, Faggionato D, O’Hara A, Kaiserli E, et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science 2011; 332:103 - 6; http://dx.doi.org/10.1126/science.1200660; PMID: 21454788
- Casati P, Walbot V. Gene expression profiling in response to ultraviolet radiation in maize genotypes with varying flavonoid content. Plant Physiol 2003; 132:1739 - 54; http://dx.doi.org/10.1104/pp.103.022871; PMID: 12913132
- Casati P, Walbot V. Differential accumulation of maysin and rhamnosylisorientin in leaves of high altitude landraces of maize after UV-B exposure. Plant Cell Environ 2005; 28:788 - 99; http://dx.doi.org/10.1111/j.1365-3040.2005.01329.x
- Casati P, Stapleton AE, Blum JE, Walbot V. Genome wide analysis of high altitude maize and gene knockdown stocks implicates chromatin remodeling proteins in responses to UV-B. Plant J 2006; 46:613 - 27; http://dx.doi.org/10.1111/j.1365-313X.2006.02721.x; PMID: 16640598
- Casati P, Walbot V. Rapid transcriptome responses of maize to UV-B in irradiated and shielded tissues. Genome Biol 2004; 5:R16; http://dx.doi.org/10.1186/gb-2004-5-3-r16; PMID: 15003119
- Casati P, Campi M, Morrow DJ, Fernandes J, Walbot V. Transcriptomic, proteomic and metabolomic analysis of UV-B signaling in maize. BMC Genomics 2011; 12:321; http://dx.doi.org/10.1186/1471-2164-12-321; PMID: 21679461
- Casati P, Campi M, Morrow DJ, Fernandes J, Walbot V. Rapid maize leaf and immature ear responses to UV-B radiation. Frontiers Plant Sci 2011; doi: 10.3389/fpls.2011.00033.
- Ulm R, Baumann A, Oravecz A, Maté Z, Adam E, Oakeley EJ, et al. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA 2004; 101:1397 - 402; http://dx.doi.org/10.1073/pnas.0308044100; PMID: 14739338
- Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, et al. A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA 2005; 102:18225 - 30; http://dx.doi.org/10.1073/pnas.0507187102; PMID: 16330762
- Oravecz A, Baumann A, Maté Z, Brzezinska A, Molinier J, Oakeley EJ, et al. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 2006; 18:1975 - 90; http://dx.doi.org/10.1105/tpc.105.040097; PMID: 16829591
- Favory J-J, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 2009; 28:591 - 601; http://dx.doi.org/10.1038/emboj.2009.4; PMID: 19165148
- Gruber H, Heijde M, Heller W, Albert A, Seidlitz HK, Ulm R. Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc Natl Acad Sci U S A 2010; 107:20132 - 7; http://dx.doi.org/10.1073/pnas.0914532107; PMID: 21041653
- Smart CC, Flores S. Overexpression of D-myo-inositol-3-phosphate synthase leads to elevated levels of inositol in Arabidopsis. Plant Mol Biol 1997; 33:811 - 20; http://dx.doi.org/10.1023/A:1005754425440; PMID: 9106505
- Loewus FA, Murthy PP. Myo-Inositol metabolism in plants. Plant Sci 2000; 150:1 - 19; http://dx.doi.org/10.1016/S0168-9452(99)00150-8
- Bohnert HJ, Nelson DE, Jensen RG. Adaptations to environmental stresses. Plant Cell 1995; 7:1099 - 111; PMID: 12242400
- Meng PH, Raynaud C, Tcherkez G, Blanchet S, Massoud K, Domenichini S, et al. Crosstalks between myo-Inositol metabolism, programmed cell death and basal immunity in Arabidopsis. PLoS ONE 2009; 4:e7364; http://dx.doi.org/10.1371/journal.pone.0007364; PMID: 19812700