Abstract
Cellular dedifferentiation is often observed in both plants and animals at an early step of wound-induced regeneration. Some plant species develop callus, a mass of unorganised cells, after wounding and this response is thought to involve cell dedifferentiation since callus cells are usually ready to exert totipotency, an ability to regenerate any new organ including somatic embryos. It is well established that a balance of the two plant hormones, auxin and cytokinin, is central in controlling plant cell dedifferentiation and subsequent redifferentiation but molecular mechanisms underlying these processes are still unclear. In a recent study we reported that an AP2/ERF transcription factor WOUND INDUCED DEDIFFERENTIATION 1 (WIND1) and its close homologs, WIND2–4, are induced by wounding and that they promote cell dedifferentiation in Arabidopsis. Our data show that WIND proteins are required to activate the local cytokinin response at the wound site.
Overexpression of WIND Homologs Induces Callus Formation and Permits Maintenance of its Dedifferentiated State
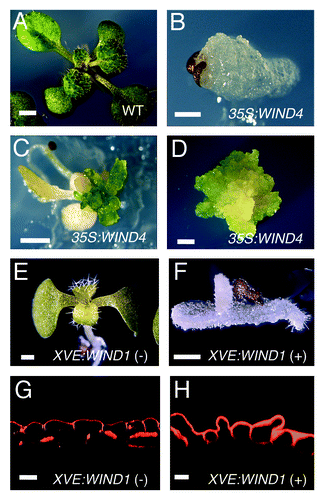
Cellular dedifferentiation, in which differentiated somatic cells become less differentiated and often start to proliferate again, occurs in many multicellular organisms after wounding.Citation1 Plant callus cells, generated at the wound site, can continue to proliferate or redifferentiate into roots, shoots, and even embryos in appropriate culture conditions.Citation2 Reacquiring these abilities is a clear sign of totipotency, strongly suggesting that callus cells are dedifferentiated. Since callus induction and subsequent alteration of their dedifferentiation/redifferentiation status are relatively easy to control by exogenous auxin and cyrokinin,Citation3 manipulation of plant cell totipotency has been widely used as a plant tissue culture technique in both basic and applied science.Citation4 Very strikingly, however, our molecular understanding of how plant cells dedifferentiate is still very limited. We have recently identified an AP2/ERF transcription factor WOUND INDUCED DEDIFFERENTIATION 1 (WIND1) and have demonstrated that WIND1 and its three other closely related homologs, WIND2, WIND3 and WIND4, participate in the regulation of cell dedifferentiation in Arabidopsis.Citation5 Our data showed that ectopic overexpression of WIND1 (At1g78080) induces typical callus formation with disorganized mass of cells on the surface of leaves, roots and hypocotyls,Citation5 and we observed similar phenotypes on transgenic plants overexpressing WIND2 (At1g22190), WIND3 (At1g36060) or WIND4 (At5g65130) under the control of Cauliflower mosaic virus 35S (35S) promoter (ref. Citation5 and ). Interestingly, callus excised from these 35S:WIND plants can proliferate on standard Murashige and Skoog (MS) medium and they have been subcultured, maintaining their dedifferentiated status, without exogenous auxin and cytokinin. To examine where callus cells originate in 35S:WIND1 plants, we induced the WIND1 expression in LexA-VP16-estrogen receptor (XVE)Citation6-WIND1 plants using 17β-estradiol (17ED) and followed the initiation of callus formation after germination. As shown in , we observed an induction of callus-like disorganized cells from the epidermal cell layer of roots, hypocotyls and cotyledons while cells in XVE-WIND1 plants, grown on control medium, continued to differentiate normally. Furthermore, we found that XVE-WIND1 plants, grown on 17ED-containing media for more than two weeks, often form somatic embryos (Iwase et al., unpublished data). Taken together, these results demonstrate that excess of WIND expression is sufficient to induce cell dedifferentiation and to maintain the dedifferentiated status in Arabidopsis.
Figure 1. Overexpression of WIND homologs promotes cell dedifferentiation without exogenous phytohormones. (A–C) 14-d-old wild-type (WT, A) and 14, 21-d-old (B, C, respectively) 35S:WIND4 seedlings grown on phytohormone-free MS medium. 35S:WIND4 seedlings display severe morphological defects and callus induction. (D) 60-d-old callus excised from 35S:WIND4 seedlings. (E, F) 8-d-old XVE-WIND1 seedlings germinated on phytohormone-free MS medium without (-) (E) or with (+) 10 mM 17b-estradiol (F). (G, H) Cross sections of cotyledon epidermal cells in 9-d-old XVE-WIND1 plants. XVE-WIND1 seedlings were germinated without (G) or with (H) 10 mM 17b-estradiol. Cellular boundaries are visualized by propidium iodide (PI) and cross sections are generated from confocal Z-stacks. Scale bars, 2 mm (A, C, D), 500 mm (B, E, F), 20 mm (G, H).
WIND1 Expression is Locally Induced at the Wound Site
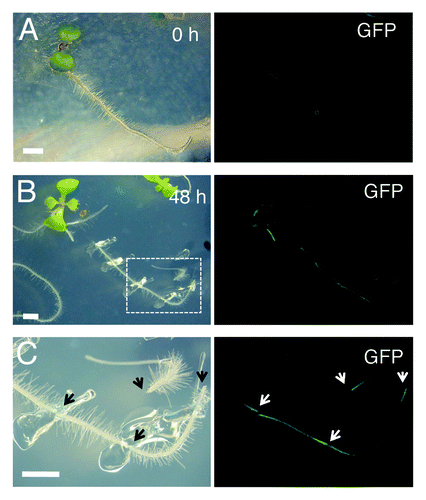
WIND1, also called RAP2.4, was listed as a wound responsive gene in previous microarray experimentsCitation7 and indeed, our quantitative RT-PCR analyses confirmed that WIND1 and all other WIND genes are rapidly upregulated within a couple of hours after wounding.Citation5 Furthermore, transgenic plants expressing the WIND1 promoter (ProWIND1)-driven reporter genes, β-glucuronidase (GUS) or green fluorescence protein (GFP) genes, showed that the WIND1 promoter activity is strongly induced at the wound site (ref. Citation5 and ). It is of particular interest that these promoter activities can be detected at both basipetal and acropetal ends of a dissected root segment (). This observation suggests that the upstream signals regulating the WIND1 promoter activity do not originate from the shoot or root apex, instead they are generated and/or activated locally at the wound site. Consistently, our quantitative RT-PCR data suggest that auxin or cytokinin, the two suspects of the apex-derived signals, do not enhance the accumulation of WIND1 transcripts.Citation5 Uncovering the signal network that translates the wound response into local WIND1 upregulation will be one of the major targets of future studies. We speculate that some of the wound-related phytohormones, such as jasmonic acids (JA), abscisic acids (ABA) and ethylene, and/or of reactive oxygen species might play a role as a transducer of the local wound response. It is to be expected that the wound-induced expression of all four WIND genes is regulated by same sets of transcription factors. Indeed the upstream promoter sequences of WIND1, 2, 3 and 4 contain several common putative cis-elements that can be recognized by transcription factors e.g., involved in JA or ABA signaling, thus it will be interesting to test whether some of these cis-elements are responsible for WIND induction at the wound site.
Figure 2. WIND1 expression is induced locally at the wound site. (Left panels) Light micrograph of 4-d-old intact ProWIND1:GFP plants (A) and 6-d-old ProWIND1:GFP plants incubated for 48 h (h) after root dissection (B). (C) Magnified view of ProWIND1:GFP root segments marked with a white box in (B). (Right panels) The WIND1 promoter activity visualized by the GFP expression. Strong GFP signals are seen at both ends of the dissected roots as indicated by arrows. Scale bars, 2 mm (A, B, C)
How do WIND Proteins Promote Cell Dedifferentiation?
Using the synthetic cytokinin reporter, two-component-output sensor (TCS)-GFP,Citation8 we have shown that the B-type ARABIDOPSIS RESPONSE REGULATOR (ARR)-mediated cytokinin response is upregulated at the wound site and that WIND proteins participate in inducing this response.Citation5 In agreement with this, 35S:WIND1 plants are hypersensitive to exogenously applied cytokinin while WIND1-SRDX (SUPERMAN repression domain)Citation9 plants, in which WIND1 function is dominantly repressed, display strongly reduced cytokinin response.Citation5 How WIND proteins promote the cytokinin signaling is not currently known but identifying direct downstream targets of the WIND-mediated transcriptional control should provide further molecular insights into how WIND proteins connect to the cytokinin signaling and/or biosynthesis pathway.
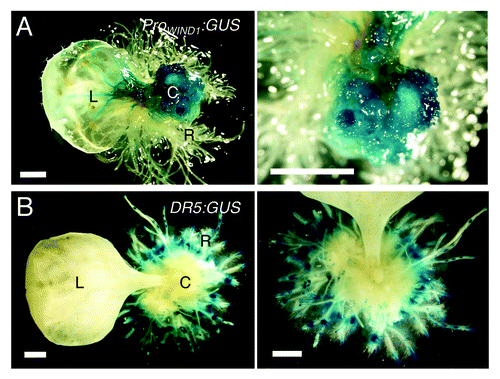
Auxin induces callus formation in many plant species including Arabidopsis and one would predict that WIND proteins also modulate auxin production or its response. Our data so far are not consistent with this view because WIND1 overexpression does not lead to increased auxin accumulation or response detectable by the cellular auxin response marker DR5:GUS.Citation10 To explore the link between auxin, WIND1 expression and callus formation more generally, we have also induced callus using synthetic auxin, 1-naphthalene acetic acid (NAA, 1 μM), in transgenic plants carrying the ProWIND1:GUS or DR5:GUS construct. As shown in , Arabidopsis leaf explants, cultured on NAA-containing medium, form callus and redifferentiate into roots extensively. As expected, the ProWIND1:GUS signal is very strong in callus cells () while interestingly, the DR5:GUS signal is hardly detectable in the equivalent callus cells (). Therefore, it appears that high WIND1 expression does not cause an increase in the auxin response at least to the level detectable by DR5:GUS and that maintaining the dedifferentiated state of callus cells does not require the level of auxin response visible by DR5:GUS.
Figure 3. WIND1 expression persists in dedifferentiated callus cells. (Left panels) Rosette leaves of 14-d-old plants carrying the ProWIND1:GUS (A) and DR5:GUS (B) constructs were cultured on 1 µM NAA containing MS medium for 30 d. (Right panels) Magnified view of the NAA-induced callus and roots. The ProWIND1:GUS signal is strong in callus cells while the DR5:GUS signal is not found in callus and detectable remarkably in roots in contact with the medium. L: leaf; C: callus; R: regenerated roots from callus. Scale bars, 1 mm (A, B)
Altogether, it is evident that WIND proteins act as a key molecular switch that links the initial wound response to the control of cell dedifferentiation. Future elucidation of how wounding upregulates the WIND transcription and how WIND mediates cell dedifferentiation should allow us advance our understanding of “how wounding promotes cell dedifferentiation”- a classic and important question in biology.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were dislosed.
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research in Innovative Areas (Grant No.22119010) and the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry
References
- Stappenbeck TS, Miyoshi H. The role of stromal stem cells in tissue regeneration and wound repair. Science 2009; 324:1666 - 9; http://dx.doi.org/10.1126/science.1172687; PMID: 19556498
- Birnbaum KD, Sánchez Alvarado A. Slicing across kingdoms: regeneration in plants and animals. Cell 2008; 132:697 - 710; http://dx.doi.org/10.1016/j.cell.2008.01.040; PMID: 18295584
- Skoog F, Miller CO. Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol 1957; 11:118 - 30; PMID: 13486467
- García-gonzáles R, Quiroz K, Carrasco B, Caligari P. Plant tissue culture: Current status, opportunities and challenges. Cienc Inv Agr 2010; 37:5 - 30
- Iwase A, Mitsuda N, Koyama T, Hiratsu K, Kojima M, Arai T, et al. The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol 2011; 21:508 - 14; http://dx.doi.org/10.1016/j.cub.2011.02.020; PMID: 21396822
- Zuo J, Niu Q-W, Chua N-H. Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 2000; 24:265 - 73; http://dx.doi.org/10.1046/j.1365-313x.2000.00868.x; PMID: 11069700
- Delessert C, Wilson IW, Van Der Straeten D, Dennis ES, Dolferus R. Spatial and temporal analysis of the local response to wounding in Arabidopsis leaves. Plant Mol Biol 2004; 55:165 - 81; http://dx.doi.org/10.1007/s11103-004-0112-7; PMID: 15604673
- Müller B, Sheen J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 2008; 453:1094 - 7; http://dx.doi.org/10.1038/nature06943; PMID: 18463635
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 2003; 34:733 - 9; http://dx.doi.org/10.1046/j.1365-313X.2003.01759.x; PMID: 12787253
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Creation of a highly active synthetic AuxRE. Plant Cell 1997; 9:1963 - 71; http://dx.doi.org/10.2307/3870557; PMID: 9401121