Abstract
The analysis of cell polarity in plants is fueled by the discovery and analysis of auxin efflux carrier PIN proteins that show polar localizations in various plant cell types in line with their roles in directional cell to cell auxin transport. As this asymmetry in cellular PIN localization drives directional auxin fluxes, abnormalities in PIN localizations modify auxin transport culminating into range of auxin distribution defective phenotypes. Because of this influence of PIN localization on plant development via changes in auxin distribution, mechanisms establishing, maintaining and altering PIN polarity are of intense interest in the plant field during the recent years. Recent findings suggest that two categories of molecules, namely AGC-3 kinase family members PINOID, WAG1, WAG2 and ARF-GEF family member GNOM predominantly influence the polar localization of PINs. The emerging mechanism for AGC-3 kinases and ARF-GEF action suggest that AGC-3 kinases predominantly phosphorylate PINs at the plasma membrane for eventual PIN internalization and PIN sorting into ARF-GEF GNOM independent polar recycling pathways. In case of mutant for AGC-3 kinases or mutations in AGC-3 kinase-targeted PIN residues, much less phosphorylated PINs are recruited into ARF-GEF GNOM-dependent polar recycling pathway. When ARF-GEF GNOM is inactive, the bias is shifted for rerouting less efficiently phosphorylated PINs into GNOM-independent polar recycling pathways that generally prefer efficiently phosphorylated PINs. Thus, balance shifts between the extent of AGC-3 kinase mediated PIN phosphorylation and the functioning of ARF-GEF instruct PIN polarity establishment and/or PIN polarity alterations. Recent studies report utilization of this AGC-3 kinase and ARF-GEF PIN polarity regulation module during diverse developmental and response programs including shoot patterning, root growth, phototropism, gravitropism, organogenesis, leaf epidermal cell indentations and fruit valve margin formation. Based on these findings the same theme of phosphorylated PIN sorting into differential polar recycling pathways for PIN polarity establishment and alteration seems to be employed in a context-dependent manner.
Breaking of cell symmetry by acquisition of polar status is a nearly universal feature of eukaryotic cells that marks the beginning of morphogenesis by providing a first visible impulse to the patterning programs. By marking distinct membrane domains cell polarity thus drives morphogenesis either in unicellular organisms such as yeast or multicellular organisms such as plants and animals. In budding yeast polarity is marked by CDC-42 protein module whereby CDC-42 loading at specific plasma membrane domain positions the site of bud outgrowth.Citation1 At many instances cell polarity in animals is marked by distinct localization of the conserved PAR protein complexes.Citation2 Whereas in extensively investigated mammalian epithelial cells physical separation of polar apical and basolateral plasma membrane domains is achieved by the membrane barrier tight junctions.Citation3 Strikingly, sequenced plants genomes do not display existence of PAR protein complexes neither do plants possess tight junctions, although some plant cell types such as mature endodermis possess membrane domain separating polymorphic barrier casparian strips.Citation4 Yet plant cells are able to acquire, maintain and modify polar status. Due to inability of plant cells to detach and move away from their neighbors as a result of shared cell walls among them modifying polarities provides the required flexibility to plant cells for switching the direction of growth in response to environmental cues and developmental programs. Thus cell polarity acquisition, maintenance and modification in plants to a large extent seems to be derived from distinct cell polarity regulation mechanisms.Citation5
The analysis of cell polarity in plants is fueled by the discovery and analysis of auxin efflux carrier PIN proteins those show asymmetric polar localization in various plant cell types in line with their roles in directional cell to cell auxin transport.Citation6–Citation8 As this asymmetry in cellular PIN localization drives directional auxin fluxes, abnormalities in PIN localizations modify directional auxin transport culminating into range of auxin distribution defective phenotypes.Citation9–Citation19 Because of this influence of PIN localization on plant development via changes in auxin distribution, mechanisms establishing, maintaining and altering PIN polarity are of intense interest during the recent years. Recent findings suggest that two category of molecules namely AGC kinase family members such as PINOID, WAG1, WAG2 and ARF-GEF family member GNOM predominantly influence the polar localization of PINs in cell type and/or context dependent manner.Citation9,Citation11–Citation14,Citation18,Citation20–Citation22 PINOID was identified as a regulator of polar auxin transportCitation23,Citation24 and later on it was shown to direct PIN localization as a binary switch in two different tissues; loss of function PID in epidermis of the shoot apical meristem results into root-ward (hereafter referred as basal) localization of PIN1 instead of its normal shootward (hereafter referred as apical) localization in WT and gain of function PID in root meristem results in apical localization of PIN1 instead of its normal basal localization in WT.Citation12 Opposite polar localization of PIN1 upon PID manipulation divert auxin flux in opposite direction resulting into defects in aerial parts in the pid mutant and defects in root growth in PID gain of function.Citation12 Recently related WAG1 and WAG2 kinases were added to the PIN polarity modifier kinase toolkit showing that WAG1 and WAG2 bear the capacity to influence PIN polarity in manner similar to that of PID and in a PID-redundant manner.Citation9 Expression analysis of PID, WAG1 and WAG2 revealed that they all are largely expressed in the cell types where PIN (e.g., PIN2 in root epidermis) shows apical localization.Citation9 Further genetic, biochemical and cell biology analysis identified the PID, WAG1 and WAG2 phosphorylation target residues present within the cytosolic loops of various PINs along with their importance for the polar PIN localization.Citation9,Citation25 Interestingly, PID, WAG1 and WAG2 phosphorylate PIN at the same residues suggesting conservation of PIN phospho-modifications by them.Citation9 Further, PIN localization pattern, PIN polarity-based polar auxin transport and auxin distribution-related plant phenotypes matched for the kinase mutants and for the manipulation of phospho-deficient PIN versions in respective PIN mutants certifying the authenticity of kinase targeted PIN phosphorylation residues. After revealing the instructive role of kinases in regulating PIN polarity and identification of the PIN residues phosphorylated by them, the next task was to uncover the cellular mechanism by which PID, WAG1 and WAG2 regulate PIN polarity. Recent results pointed that all the three kinases localize at the plasma membrane in a non-polar manner for impacting PIN polarization.Citation9 Detailed time lapse analysis revealed that after synthesis PID, WAG1 and WAG2 load at the plasma membrane in a membrane sterol composition dependent manner to phosphorylate PINs.Citation9 This occurs when PINs arrive at the plasma membrane for the first time after their non-polar secretion or when PINs return to the plasma membrane after undergoing a cycle of endocytosis and recycling.Citation9 Together these results put forward a model whereby PID, WAG1 and WAG2 phosphorylate PINs largely at the plasma membrane after default non-polar PIN secretion and thus provide a phosphorylation signature to the PIN.Citation9 This PIN modification presumably is then used for their post-endocytic sorting at the intermediate compartments located along the trafficking route to eventually recruit them in differential recycling pathways for their deposition at a particular cell side.Citation9 It was also found that even with the conserved phosphorylation residues different PINs display different degree of sensitivity for their kinase-triggered polar localization.Citation9,Citation25 This occurs in a PIN sequence- and cell type-dependent manner suggesting that the sequenced-based intrinsic differences within different PINs and the cell type-based extrinsic differences both influence the PIN behavior.Citation9 Thus identification of PIN phosphorylation residues along with isolation of the cellular mechanism for kinase action on PIN polarity offer promising possibilities to manipulate PIN polarity in various contexts to further probe the contribution of PIN polarity in various plant developmental and response programs.
An ARF-GEF GNOM was identified as general polarity regulator in genetic screen for plant embryo patterning mutants whereby gnom displayed embryonic defects such as fused cotyledons and formation of residual or no root.Citation15,Citation26,Citation27 Further studies identified PIN1 as the substrate of GNOM and determined predominant localization of GNOM at the recycling endosomes implicating GNOM in polar endocytic recycling of PIN1.Citation15,Citation20 It was also shown that a drug BFA inactivates GNOM impairing GNOM-dependent PIN1 recycling that results into massive aggregation of PIN1 positive vesicles to form large cytosolic BFA compartments.Citation15,Citation20,Citation28 Once GNOM was engineered to become BFA insensitive the GNOM dependent PIN1 recycling, PIN1 polarity and PIN1 mediated auxin distribution remained intact in presence of BFA.Citation20 In a root meristematic epidermal cell PIN1 remains at the basal cell side whereas PIN2 at the apical cell side and in a root meristematic cortical cell both PIN1 and PIN2 remain at the basal cell side.Citation29–Citation31 Strikingly, prolonged BFA treatments gradually shifted the basally localized PIN1 in epidermis and basally localized PIN1 and PIN2 in cortex towards the apical cell sides via an intermediate BFA compartment stopover in process termed as transcytosis.Citation13 This is considered to occur via PIN1 and PIN2 endocytosis from the basal cell side for their aggregation into BFA compartments (via a BFA sensitive ARF-GEF such as GNOM) followed by BFA compartment to apical cell side translocation of PIN1 and PIN2 (via a BFA resistant ARF-GEF).Citation13 Interestingly, PID, WAG1 and WAG2 induction in those cell types displayed similar PIN translocation from basal to apical cell side with an intact GNOM functioning and without intermediate accumulation of PINs in the BFA compartments.Citation9,Citation21 Further analysis revealed that (1) basal to apical translocation of PINs in BFA treatment-induced GNOM inactive situation depends on PID-, WAG1-, WAG2-mediated PIN phosphorylation (2) phospho-deficient PINs are unable to achieve clear apical localization in case of BFA treatment-induced GNOM inactive situation and (3) post PIN apicalization induction stop of PID, WAG1, WAG2 by removal of induction chemical leads to apical to basal PIN translocation via GNOM regulated basal recycling pathway.Citation9,Citation21 Together these findings suggested that PID and GNOM act antagonistically.
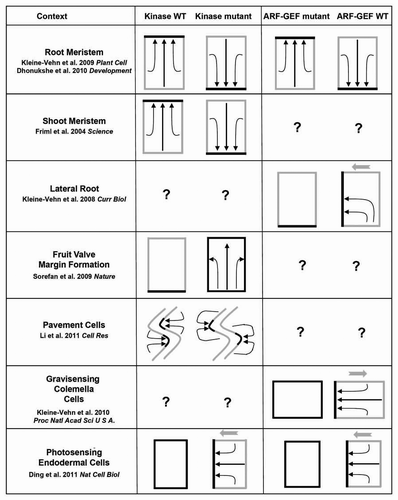
The same theme of kinase phosphorylated PIN sorting for incorporation into ARF-GEF based differential polar recycling pathway for eventual PIN polarity establishment or PIN polarity alteration seems to be employed in a context-dependent manner. Recent studies report utilization of conserved PID-GNOM (AGC-3 kinase and ARF-GEF) module for PIN polarity alteration during diverse developmental and response programs including phototropism,Citation11 gravitropism,Citation14 organogenesis such as lateral root formation,Citation13 leaf epidermal cell indentationsCitation18 and fruit valve margin formationCitation22 (see ). These diverse developmental and response programs either use different PINs or use modified versions of PID-GNOM module whereby PINs display non-polar to one cell side, one cell side to another cell side or polar to less polar translocation (). At some instances the antagonistic nature of PID-GNOM pair is demonstrated while at other instances influence of either PID alone or GNOM alone has been implicated in PIN polarity alterations (). Light- and auxin-regulated PID transcription adds another layer of controlCitation11,Citation24 for PID mediated PIN polarity regulation events. As evident in there remains some questions those need further investigation to validate the universality of PID-GNOM antagonistic action concept.
Decade of intense research on PIN polarity yet leaves unsolved mystery of how the vesicle traffic pathway(s) involving PID-GNOM antagonism provide a directional impulse for regulating deposition and maintenance of PIN at one particular cell side. PINs display less lateral diffusion within the plant plasma membrane as compared to other non-polar cargoes in both polar cells (where PINs are polar) within the tissueCitation10 or in single cultured non-polar cells (where PINs are non-polar) such as cell wall deficient protoplastsCitation32 removing the possibility that less lateral diffusion is instructive in regulating PIN polarity. Polar endocytosis by localized removal of PINs from areas where PIN is not needed seems also a less favorable option for PIN polarity attainment especially when the non-polar cargoes also utilize the very same endocytotic pathway that PINs use.Citation32 With this many unanswered questions as mentioned below still pose hypothetical options. Is it that all cell sides bear equal capacities to retain PINs and there are cell side-oriented specialized endocytic recycling pathways? Is it that the directionality influenced selective spatiotemporal dominance of one particular cell side-oriented endocytic recycling pathway provides the bias for gradual buildup of PIN on one particular cell side? Is it that the cells are under constant dialogue with their neighbors and just decide to switch their intrinsic cell side identities and cell side identity-associated endocytic recycling pathway to reposition PINs? Is it that the spatiotemporal polarization of cytoskeleton provides the bias for mass delivery of PIN positive vesicles to one particular cell side? Another unanswered fact is how the common vesicle trafficking pathways distinguish between the polar and non-polar cargoes especially when both the polar and non-polar cargoes reside transiently in the very same intermediate trafficking compartments? Is it that the same vesicles carry different cargoes by allocating distinct vesicular domains to different cargoes or is it that distinct vesicles are pinched off from the common intermediate compartment for selective transfer of polar and non polar cargoes? Further, feedback nature of auxin and PIN relation via influence of intra- and/or extra-cellular auxin on clathrin mediated endocytic pathway which is used by both PINs and non-polar cargoesCitation32–Citation34 and via PIN regulated abundance of intra- and/or extra-cellular auxin certainly complicate the unraveling of any potential hierarchy in PIN polarity module. Recent reports also put forward the influence of stress and strain patterns on PIN polarity via mechanical signalingCitation35,Citation36 which presumably is in a constant dialogue with the biochemical gene transcriptional and protein modifications programs. In addition, the reports from non-plant field revealing that polarity regulators control membrane trafficCitation37 and localization of polarity regulators is directed by membrane trafficCitation38 add more to the complexity of the polarity puzzle. Even though the coarse mechanism of PIN polarity generation from non-polar secretion followed by PID-GNOM competition based polar endocytic recycling seems to be solvedCitation9,Citation10 there is yet much to learn about the ways plants cells establish, maintain and alter PIN polarity. How PINs are destined to one particular cell side during typical developmental or response program certainly remains a central question with a big challenge to solve the issue of directionality. With the availability of exciting in vivo traceable tools such as photoconvertible fluorescent tag attached functional PIN versions, progressive and high resolution microscopy platforms, constant identification of new PIN trafficking regulators and acquisition of finite details of the AGC-3 kinase and ARF-GEF PIN sorting module should bring us closer towards the answers.
Figures and Tables
Figure 1 Roles of PID-GNOM (AGC-3 kinase and ARF-GEF) module for instructing polar PIN localization during plant developmental and response programs. The gray rectangles depict plant cells and the black sides of the gray rectangles show PIN localization. The black arrows within the gray rectangles show translocation of PINs from one cell side to another. The gray arrows indicate the direction of the gravity impulse or the direction of the light impulse or the direction of the lateral root outgrowth.
Acknowledgments
P.D. acknowledges the valuable contributions of plant cell polarity research community those made materialization of this review possible and apologizes to those whose work could not be cited due to space constraints. P.D. is thankful to Jiri Friml, Ben Scheres, Remko Offringa and Zenbiao Yang for the past and current fruitful discussions related to the polarity topic. P.D. is supported by the Dutch Scientific Organization's VENI grant and by the Utrecht University Starting Independent Investigator grant.
References
- Etienne-Manneville S. Cdc42—the centre of polarity. J Cell Sci 2004; 117:1291 - 1300
- Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev Cell 2007; 13:609 - 622
- Shin K, Fogg VC, Margolis B. Tight junctions and cell polarity. Annu Rev Cell Dev Biol 2006; 22:207 - 235
- Roppolo D, De Rybel B, Tendon VD, Pfister A, Alassimone J, Vermeer JE, et al. A novel protein family mediates Casparian strip formation in the endodermis. Nature 473:380 - 383
- Geldner N. Cell polarity in plants: a PARspective on PINs. Curr Opin Plant Biol 2009; 12:42 - 48
- Dhonukshe P. Cell polarity in plants: Linking PIN polarity generation mechanisms to morphogenic auxin gradients. Commun Integr Biol 2009; 2:184 - 190
- Galweiler L, Guan C, Muller A, Wisman E, Mendgen K, Yephremov A, et al. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 1998; 282:2226 - 2230
- Grunewald W, Friml J. The march of the PINs: developmental plasticity by dynamic polar targeting in plant cells. EMBO J 29:2700 - 2714
- Dhonukshe P, Huang F, Galvan-Ampudia CS, Mahonen AP, Kleine-Vehn J, Xu J, et al. Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 137:3245 - 3255
- Dhonukshe P, Tanaka H, Goh T, Ebine K, Mahonen AP, Prasad K, et al. Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions. Nature 2008; 456:962 - 966
- Ding Z, Galvan-Ampudia CS, Demarsy E, Langowski L, Kleine-Vehn J, Fan Y, et al. Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat Cell Biol 13:447 - 452
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, et al. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 2004; 306:862 - 865
- Kleine-Vehn J, Dhonukshe P, Sauer M, Brewer PB, Wisniewska J, Paciorek T, et al. ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Curr Biol 2008; 18:526 - 531
- Kleine-Vehn J, Ding Z, Jones AR, Tasaka M, Morita MT, Friml J. Gravity-induced PIN transcytosis for polarization of auxin fluxes in gravity-sensing root cells. Proc Natl Acad Sci USA 107:22344 - 22349
- Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S, et al. Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 1999; 286:316 - 318
- Furutani M, Sakamoto N, Yoshida S, Kajiwara T, Robert HS, Friml J, et al. Polar-localized NPH3-like proteins regulate polarity and endocytosis of PIN-FORMED auxin efflux carriers. Development 138:2069 - 2078
- Hazak O, Bloch D, Poraty L, Sternberg H, Zhang J, Friml J, et al. A rho scaffold integrates the secretory system with feedback mechanisms in regulation of auxin distribution. PLoS Biol 8:1000282
- Li H, Lin D, Dhonukshe P, Nagawa S, Chen D, Friml J, et al. Phosphorylation switch modulates the interdigitated pattern of PIN1 localization and cell expansion in Arabidopsis leaf epidermis. Cell Res 21:970 - 978
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 2007; 130:1044 - 1056
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, et al. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport and auxin-dependent plant growth. Cell 2003; 112:219 - 230
- Kleine-Vehn J, Huang F, Naramoto S, Zhang J, Michniewicz M, Offringa R, et al. PIN auxin efflux carrier polarity is regulated by PINOID kinase-mediated recruitment into GNOM-independent trafficking in Arabidopsis. Plant Cell 2009; 21:3839 - 3849
- Sorefan K, Girin T, Liljegren SJ, Ljung K, Robles P, Galvan-Ampudia CS, et al. A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature 2009; 459:583 - 586
- Christensen SK, Dagenais N, Chory J, Weigel D. Regulation of auxin response by the protein kinase PINOID. Cell 2000; 100:469 - 478
- Benjamins R, Quint A, Weijers D, Hooykaas P, Offringa R. The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 2001; 128:4057 - 4067
- Huang F, Zago MK, Abas L, van Marion A, Galvan-Ampudia CS, Offringa R. Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 22:1129 - 1142
- Shevell DE, Leu WM, Gillmor CS, Xia G, Feldmann KA, Chua NH. EMB30 is essential for normal cell division, cell expansion and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7. Cell 1994; 77:1051 - 1062
- Geldner N, Richter S, Vieten A, Marquardt S, Torres-Ruiz RA, Mayer U, et al. Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development 2004; 131:389 - 400
- Geldner N, Friml J, Stierhof YD, Jurgens G, Palme K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 2001; 413:425 - 428
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, et al. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005; 433:39 - 44
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev 1998; 12:2175 - 2187
- Wisniewska J, Xu J, Seifertova D, Brewer PB, Ruzicka K, Blilou I, et al. Polar PIN localization directs auxin flow in plants. Science 2006; 312:883
- Dhonukshe P, Aniento F, Hwang I, Robinson DG, Mravec J, Stierhof YD, et al. Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol 2007; 17:520 - 527
- Paciorek T, Zazimalova E, Ruthardt N, Petrasek J, Stierhof YD, Kleine-Vehn J, et al. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 2005; 435:1251 - 1256
- Robert S, Kleine-Vehn J, Barbez E, Sauer M, Paciorek T, Baster P, et al. ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell 143:111 - 121
- Hamant O, Heisler MG, Jonsson H, Krupinski P, Uyttewaal M, Bokov P, et al. Developmental patterning by mechanical signals in Arabidopsis. Science 2008; 322:1650 - 1655
- Heisler MG, Hamant O, Krupinski P, Uyttewaal M, Ohno C, Jonsson H, et al. Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PLoS Biol 8:1000516
- Balklava Z, Pant S, Fares H, Grant BD. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat Cell Biol 2007; 9:1066 - 1073
- Osmani N, Peglion F, Chavrier P, Etienne-Manneville S. Cdc42 localization and cell polarity depend on membrane traffic. J Cell Biol 191:1261 - 1269