Abstract
Maturation of the 40S ribosomal subunit precursors in mammals mobilizes several non-ribosomal proteins, including the atypical protein kinase RioK2. Here, we have investigated the involvement of another member of the RIO kinase family, RioK3, in human ribosome biogenesis. RioK3 is a cytoplasmic protein that does not seem to shuttle between nucleus and cytoplasm via a Crm1-dependent mechanism as does RioK2 and which sediments with cytoplasmic 40S ribosomal particles in a sucrose gradient. When the small ribosomal subunit biogenesis is impaired by depletion of either rpS15, rpS19 or RioK2, a concomitant decrease in the amount of RioK3 is observed. Surprisingly, we observed a dramatic and specific increase in the levels of RioK3 when the biogenesis of the large ribosomal subunit is impaired. A fraction of RioK3 is associated with the non ribosomal pre-40S particle components hLtv1 and hEnp1 as well as with the 18S-E pre-rRNA indicating that it belongs to a bona fide cytoplasmic pre-40S particle. Finally, RioK3 depletion leads to an increase in the levels of the 21S rRNA precursor in the 18S rRNA production pathway. Altogether, our results strongly suggest that RioK3 is a novel cytoplasmic component of pre-40S pre-ribosomal particle(s) in human cells, required for normal processing of the 21S pre-rRNA.
Introduction
While the nascent pre-rRNA is being transcribed by RNA polymerase I in the nucleolus, it associates with a subset of snoRNPs, ribosomal and non-ribosomal proteins, resulting in the assembly of so-called pre-ribosomal particles.Citation1-Citation6 Either during transcription or post-transcriptionally, the newly transcribed pre-rRNA undergoes endonucleolytic cleavages that release the precursors to the large and small ribosomal subunits, the pre-60S and pre-40S particles respectively.Citation7 These precursors will then undergo separate and largely independent maturation pathways. In the course of their maturation, pre-ribosomal particles transit from the nucleolus to the cytoplasm via the nucleoplasm and nuclear poresCitation4,Citation8,Citation9. Both types of precursor particles undergo final maturation steps in the cytoplasm to acquire translational competence (for reviews see refs. Citation8 and Citation10).
More than a hundred of non-ribosomal proteins dynamically associate with and dissociate from pre-ribosomal particles at various stages of their maturation.Citation2,Citation10,Citation11 Among these non-ribosomal proteins feature endo- and exoribonucleases, nucleotide modifying enzymes, RNA helicases, NTPases, kinases, RNA-binding proteins etc… However, many non-ribosomal proteins fail to display any easily recognizable motif. Most of these factors are essential for viability and ribosome synthesis in yeast, yet their precise molecular functions remain elusive, with the notable exception of nucleases involved in pre-rRNA processing. These non-ribosomal proteins may promote ribosomal protein assembly, pre-rRNA structural rearrangements, pre-rRNA spacer digestion, pre-ribosomal particle transport or participate in quality control of ribosome biogenesis.
Maturation of pre-40S pre-ribosomal particles may prove less complex than that of pre-60S particles, as far fewer non-ribosomal proteins associate with pre-40S particles, at least in yeast.Citation12 The major non-ribosomal constituents of yeast pre-40S particles are the kinases Hrr25p,Citation13 Rio1p and Rio2p,Citation14,Citation15 the 18S rRNA methyl-transferase Dim1p,Citation16,Citation17 the Nob1p endonuclease,Citation18-Citation20 the putative RNA-binding protein Dim2p,Citation21,Citation22 the heat repeat protein Rrp12pCitation23 and the Enp1p,Citation24,Citation25 Ltv1pCitation3,Citation26 and Tsr1pCitation27 proteins. In addition, pre-40S particles transiently interact with the Fap7p NTPaseCitation28 and with the DEAH helicase Prp43p in complex with its G-patch-containing protein partner Pfa1p.Citation20,Citation29 Dim1p, Dim2p, Enp1p, Hrr25p, Nob1p, Rrp12p and Tsr1p probably associate early with nucleolar 90S pre-ribosomal particles,Citation3,Citation30 while Ltv1p, Rio1p and Rio2p join later nuclear pre-40S particles generated following pre-RNA cleavage at site A2 in internal transcribed spacer 1.Citation31,Citation32 The molecular functions of some of these factors are starting being defined. Nob1p is the endonuclease that converts yeast 20S pre-rRNA into 18S rRNA in the cytoplasm,Citation20,Citation33,Citation34 following a conformational rearrangement of cytoplasmic pre-40S particles requiring Prp43p, Pfa1p and Ltv1p. The Hrr25p kinase is responsible for the nuclear phosphorylation of ribosomal protein Rps3p and its direct partner Enp1p, which weakens the association of Rps3p and Enp1p with the pre-40S particles, a likely prerequisite for passage of these particles through the nuclear pore.Citation30 The two other kinases present in yeast pre-40S particles, the Rio1p and Rio2p kinases, are each required for the cytoplasmic conversion of 20S pre-rRNA into 18S rRNA, demonstrating that they do not play redundant functions in ribosome biogenesis.Citation14,Citation15 The composition of human pre-40S particles is starting being defined. So far, it appears that human and yeast pre-40S particles share several highly conserved non-ribosomal protein components, including hEnp1/bystin, hLtv1 and RioK2.Citation35 However, the pre-40S maturation pathway is more complex in human than in yeast cells: elimination of the ITS1-fragment within the yeast pre-40S particles requires a single cytoplasmic cleavage step at the 3′ end of the 18S rRNA, while one detects 3 intermediates in pre-40S particles in human cells, namely the 21SCitation36 and 21S-CCitation37 pre-rRNAs that are nuclear, and the cytoplamic 18S-E pre-rRNA.Citation38
The yeast Rio1p and Rio2p and the human RioK2 proteins are members of the RIO (right open reading frame) family of atypical protein kinases (aPK).Citation39 This family is characterized by the presence of the RIO kinase domain, a kinase fold structurally homologous to eukaryotic protein kinase (ePK) domains but lacking the loops known to be important for substrate binding in the canonical kinases, as well as the so-called “activation loop,” the phosphorylation of which modulates kinase activity.Citation40-Citation42 Another distinguishing feature of the RIO kinase domain is the presence of a “flexible loop” that may interfere with nucleotide binding.Citation43 The Rio1 and Rio2 proteins are conserved from archaea to mammals. In contrast, some eubacterial species contain a single representative of the RIO protein family, displaying similarities to Rio1 and Rio2 proteins at its N- and C-terminus, respectively, suggesting that a common ancestor of the two proteins may have existed.Citation44,Citation45 A third type of RIO protein named Rio3 was described, which is more similar to Rio1.Citation39 Genes encoding Rio3 proteins have only been found in multicellular eukaryotes thus far. In humans, the three RIO kinases have been termed RioK1, RioK2 and RioK3. RioK2 has been shown to be required for the conversion of the 18S-E pre-rRNA precursor into mature 18S rRNA, confirming the requirement for Rio2 proteins in the last step of mature 18S rRNA production,Citation38 and recycling of several non-ribosomal factors.Citation35 In the present study, we have investigated the role of the human RioK3 protein in ribosome biogenesis. Our results strongly suggest that RioK3 is a component of cytoplasmic pre-40S particles and show that its depletion leads to an accumulation of the 21S pre-rRNA, a precursor to 18S rRNA. Hence human 40S ribosomal subunit biogenesis mobilizes an additional member of the RIO kinase family compared with yeast.
Results
Human RioK3 is cytoplasmic and sediments with 40S ribosomal particles
The RioK3 protein is highly conserved among multicellular eukaryotes (). The RioK3 central domain is a canonical RIO domain, based on the founding member of the family, yeast Rio1p, containing an ATP-binding site, a flexible loop, a hinge, a catalytic site and a metal- binding loop.Citation46 The RioK3 primary structure also features a conserved N-terminal domain (1–255) predicted to be highly helical, but displaying no sequence homology to any known protein (). At the beginning of our study, the RioK3 protein was totally uncharacterized experimentally. We thus first determined its sub-cellular localization. HeLa cells expressing a cDNA encoding this protein fused to the enhanced green fluorescent protein (EGFP) displayed a fluorescent signal restricted to the cytoplasm when observed by confocal microscopy (). Consistent with this observation, after cellular fractionation, endogenous RioK3 detected by western blot analysis was only found in the cytoplasmic fraction (). To assess whether RioK3 enters the nucleus and is rapidly exported back in the cytoplasm by a Crm1-dependent mechanism, cells expressing EGFP-RioK3 were treated for 2 h with Leptomycin B (LMB), a drug inhibiting the exportin Crm1.Citation47 Most of EGFP-RioK3 remained in the cytoplasm (), when under similar conditions, EGFP-RioK2 became concentrated in the nucleus (), as previously described for both yeast Rio2p and human RioK2.Citation15,Citation38 These results suggest that, contrary to these latter proteins, RioK3 is not actively exported from the nucleus by a Crm1-dependent mechanism, at least in the HeLa cells we are using. Moreover, should RioK3 be actively exported from the nucleus, this probably does not occur by “piggybacking” on pre-ribosomal particles as inhibition of ribosome biogenesis by actinomycin D did not perturb EGFP-RioK3 localization (data not shown).
Figure 1. RioK3 conservation, sub-cellular localization and sedimentation on density gradient. (A) Alignment of RioK3 from diverse organisms (X. laevis, E. caballus, M. musculus, B. taurus, M. fascicularis and H. sapiens) showing the high degree of conservation of the protein. The conserved N-ter domain is framed in blue. The RIO kinase domain contains the ATP-binding loop, the flexible loop, the hinge region, and the catalytic and metal-binding loops as determined from the structure of the A. fulgidus Rio2 protein.Citation55,Citation56 (B) Schematic diagram of the domain organization of RioK3 showing sequence signatures within the rectangle representing the RIO domain. The well conserved N and C domains have no sequence homologs in any known protein. (C) HeLa cells expressing EGFP-RioK3 and EGFP observed by laser-scanning confocal microscopy. (D) western blot analysis of HeLa cell cytoplasmic and nuclear fractions performed with an anti-RioK3 antibody showing that RioK3 is a cytoplasmic protein. As a control, the component of the box HACA snoRNPs dyskerin is detected mainly in the nuclear fraction. (E) 48 h after transfection with the EGFP-RioK3 (left) or EGFP-RioK2 (right) expression vector, cells were treated with 10 nM LMB for 2 h and then observed by laser-scanning confocal microscopy. EGFP-RioK3 localization is not affected when Crm1 is inhibited by LMB, in contrast to EGFP-RioK2 which is clearly retained in the nucleus. (F) Centrifugation of a HeLa cell cytoplasmic extract on a 15–50% sucrose gradient and Western/Northern blot analyses of the gradient fractions. Fractions were analyzed by western blot with antibodies against RioK3, RioK2 and Rps19. The 18S and 28S rRNAs were detected by ethidium bromide staining and the 18S-E species was revealed by northern blotting with a 5′-ITS1 probe. This analysis shows that the totality of RioK3 co-sediments with (pre)-40S particles, as does RioK2.
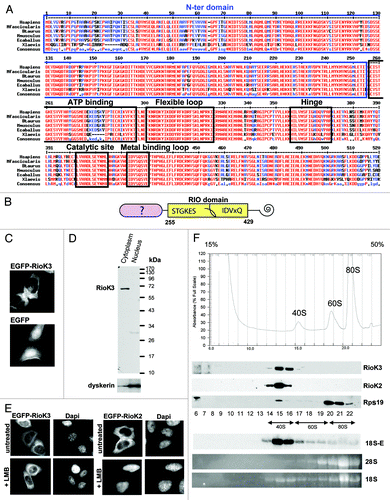
We next assessed the sedimentation profile of RioK3 on a sucrose density gradient. A cytoplasmic fraction prepared in the presence of cycloheximide was separated on a 15–50% (w/v) sucrose density gradient (). The distribution of free 40S and 60S ribosomal subunits and 80S ribosomes was assessed by monitoring absorbance at 254 nm, and by detection of the 18S and 28S rRNAs in each fraction (note that on this type of gradient, polysomes are sedimenting at the bottom of the tube and are absent from the profile). Fractions containing pre-40S particles were identified by the presence of the 18S-E pre-rRNA detected by northern-blotting with a 5′-ITS1 probe. The sedimentation on sucrose gradients of RioK3 and RioK2 as control was analyzed by the western blotting procedure using specific antibodies. The totality of RioK3 sediments with free 40S/pre-40S ribosomal subunits, as does RioK2. Altogether, these results suggest that RioK3 could be a component of cytoplasmic pre-40S particles or of the cytoplasmic 40S ribosomal subunit.
RioK3 levels are reduced when the small ribosomal subunit (SSU) biogenesis is impaired
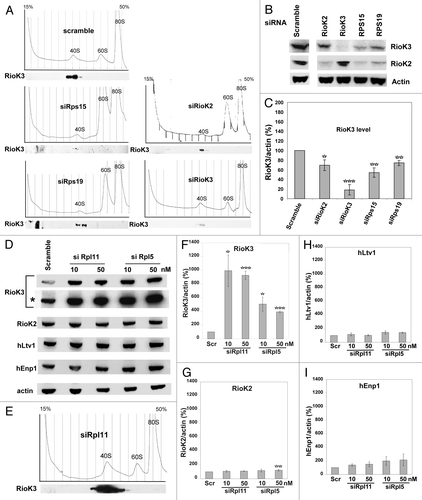
In order to determine whether RioK3 belongs to some pre-40S particle(s), we analyzed the effect of SSU biogenesis impairment on the levels of RioK3. As shown in , depletion of the small subunit ribosomal proteins Rps15 or Rps19 using appropriate siRNAs resulted, as expected, in a significant drop in the levels of free 40S particles, with a concomitant increase in free 60S particles. Under these conditions, RioK3 sedimentation profile was unchanged: it was still present in the gradient fractions containing the remaining free 40S particles but its amount was decreased by approximately 50% or 30% in the case of Rps15 or Rps19 depletion, respectively (). Likewise, depletion of Rps15 or Rps19 significantly decreased the levels of RioK2 (). Moreover, knockdown of RioK2 expression by siRNA treatment also resulted in a significant drop in 40S particle levels correlated with an approximate 30% decrease in RioK3 levels (). Together, these results indicate that the amount of RioK3 vary with the levels of free 40S particles or pre-40S particles, as is the case for RioK2, suggesting that RioK3 may be a component of pre-40S particles or 40S subunits.
Figure 2. Impact of Rps15, Rps19, RioK2, Rpl5 or Rpl11 depletion on ribosome biogenesis and accumulation of RioK3 and other components of pre-40S particles. (A) Cytoplasmic extracts, prepared from cycloheximide-treated cells 48h after transfection with a scramble siRNA or a siRNA targeting Rps15, Rps19, RioK2 or RioK3, were subjected to ultracentrifugation in 15–50% sucrose gradients. Identical amounts of each fraction were analyzed by western using an anti-RioK3 antibody. Detection of the chemiluminescence by autoradiography was performed for the same duration in all cases. (B) Accumulation of RioK3 and RioK2 in siRNA-treated cells described in (A). Total cellular extracts from these cells were analyzed by western using antibodies against RioK3, RioK2 and actin. (C) Quantifications of the amounts of RioK3 performed using a Bioimager after western blot analysis. The quantity of each protein was normalized to that of actin and expressed as percentages, the quantity in cells treated with scramble siRNAs being set at 100%. Values presented are the means (+/−SEM) of 5 independent experiments. Pair-wise statistical analysis was performed with Student’s t-test. *p < 0.05; **p < 0.01; ***p < 0.001. (D) Total cellular extracts prepared 48 h after transfection with either scramble, rpL11 (10 and 50 nM) or rpL5 (10 and 50 nM) siRNAs were analyzed by the Western procedure with antibodies specific for RioK3, RioK2, hLtv1, hEnp1 and actin. A longer exposure (*) is shown in the case of RioK3. (E) Sucrose gradient sedimentation profile of ribosomal particles from a cytoplasmic extract produced from siRpl11-treated cells. Fractions were analyzed by the Western procedure with an anti-RioK3 antibody. Blots were exposed for the same time in (D) and (E). (F-I) Quantifications of the amounts of RioK3 (F), RioK2 (G), hLtv1 (H), hEnp1 (I) performed using a Bioimager after western blot analysis. As in (C), the quantity of each protein was normalized to that of actin and expressed as percentages, the quantity in cells treated with scramble siRNAs being set at 100%. Values presented are the means of 3 independent experiments (+/−SEM). Pair-wise statistical analysis was performed with Student’s t-test. *p < 0.05; **p < 0.01; ***p < 0.001.
RioK3 levels are specifically increased when large ribosomal subunit (LSU) biogenesis is impaired
We then assessed the consequence of inhibition of LSU biogenesis on RioK3 levels. Strikingly, when LSU biogenesis was impaired in HeLa cells by depletion of either Rpl11 or Rpl5 using siRNAs, we observed a dramatic and specific increase (approximately 10-fold) in RioK3 levels () as compared with scramble siRNA-treated cells. In extracts from siRpl5-treated cells, the increase in RioK3 levels was lower (~5 fold increase) than that observed in siRpl11-treated cells (). We also detected a significant and specific increase (~5-fold) in RioK3 levels in U2OS cells transfected with either Rpl11 or Rpl5 siRNAs (data not shown). Analysis of the ribosomal profile on sucrose gradient after Rpl11 depletion confirmed that the amount of free 60S particles was significantly decreased, with a concomitant increase in free 40S particles as expected for the depletion of a LSU ribosomal protein () as compared with the profile obtained with extracts from scramble siRNA-treated cells (). All RioK3 was found in the 40S fractions of the gradient, suggesting that even when the amount of RioK3 is vastly increased, most RioK3 proteins remain associated with the pre-40S particles and/or 40S ribosomal subunits. We then checked whether the levels of other pre-40S factors were also increased when LSU biogenesis was impaired. As previously described for RioK3, the amounts of RioK2, hLtv1 and hEnp1 were determined by western blot analysis using cellular extracts from scramble, siRpl11 or siRpl5-treated cells (). In extracts from siRpl11-treated cells, the levels of RioK2, hLtv1 and hEnp1 remained fairly constant or show a slight increase (1.1- to 2.4-fold, ). Altogether, these results confirm the correlation between the levels of RioK3 and those of free 40S/pre-40S particles.
RioK3 is associated with pre-40S particle components hLtv1, hEnp1 and 18S-E
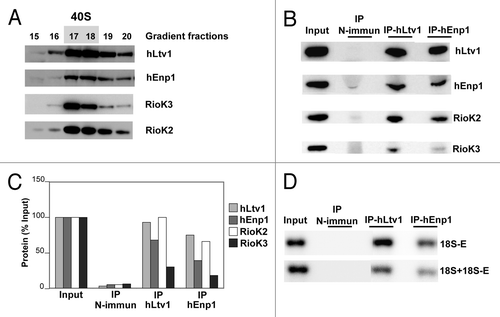
We next sought to directly assess whether RioK3 is associated with pre-40S particle components by immunoprecipitation. We failed to efficiently precipitate endogenous RioK3 using the commercially available anti-RioK3 antibody (not shown). Therefore, we decided to check whether RioK3 is co-precipitated with protein components of human pre-40S particles. We made use of good antibodies against hEnp1/bystin and hLtv1, known components of human pre-40S particles.Citation35,Citation37 A HeLa cytoplasmic extract was fractionated on a preparative sucrose gradient and the 40S fractions containing hLtv1, hEnp1, RioK2 and RioK3 were pooled () and subjected to immunoprecipitation with anti-hLtv1 or anti-hEnp1 antibodies. The anti-hLtv1 antibodies immunoprecipitated hLtv1 very efficiently (). Use of these antibodies also led to the very efficient co-precipitation of hEnp1 and RioK2 (approximately ~70–95% recovery), while RioK3 was also co-precipitated, but to a lesser extent (approximately ~30% recovery, ). Similarly, the anti-hEnp1 antibodies immunoprecipitated efficiently hEnp1 together with hLtv1, RioK2 and to a far lesser extent RioK3 (). Northern blot analysis of the RNAs co-precipitated with anti-hLtv1 or anti-hEnp1 antibodies showed that the 18S-E pre-rRNA was recovered with a very high yield (respectively 90% and 56% of input) (). Hybridization with a probe complementary to 18S rRNA (and necessarily to 18S-E pre-rRNA also) shows that 35% and 20% of the combined 18S rRNA + 18S-E pre-rRNA input species are precipitated with hLtv1 and hEnp1, respectively. These results strongly suggest that RioK3 is a component of a particle containing also two bona fide components of pre-40S particles and 18S-E pre-rRNA.
Figure 3. Immunoprecipitation of pre-40S particles. (A) A HeLa cell cytoplasmic extract was subjected to ultracentrifugation in a preparative sucrose gradient. Gradient fractions were analyzed by the western blot procedure with antibodies against hLtv1, hEnp1, RioK3 and RioK2. Fractions 17 and 18, containing the bulk of these proteins and presumably pre-40S particles, were pooled. (B) western blot analysis of proteins precipitated from these fractions using IgG beads pre-incubated with non-immune (control), anti-hLtv1 or anti-hEnp1 sera. Proteins were extracted from the bead pellets (75% of the IP samples loaded) or from a volume of gradient fractions equivalent to that used as input (Input) and analyzed by western blot with antibodies against hLtv1, hEnp1, RioK3 and RioK2. (C) Percentage of protein recovery following immunoprecipitation. (D) northern blot analysis of the co-precipitated RNAs. RNAs loaded were extracted from 30 µl of the input 40S fraction (Input) or from the bead pellets (IP) obtained following immunoprecipitation performed with the same fraction volume and analyzed by the northern procedure using the 5′-ITS1 probe to detect 18S-E pre-rRNA, and the 18S probe to detect 18S rRNA + 18S-E pre-rRNA (see Table S1).
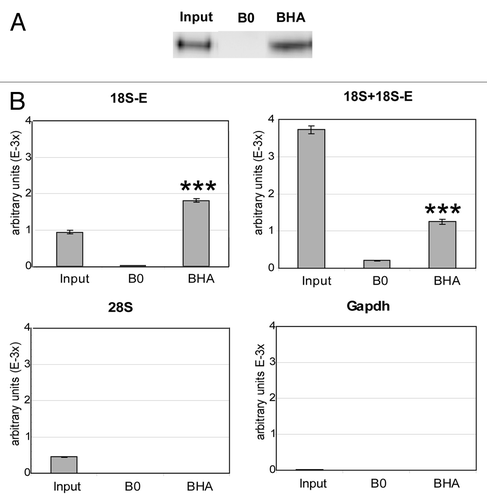
To confirm that RioK3 itself is associated with 18S-E pre-rRNA, we performed a direct immunoprecipitation of a transiently expressed HA-tagged RioK3. A cytoplasmic extract from HeLa cells transfected with plasmid E46-HA-RioK3 allowing the transient expression of HA-RioK3 (that co-sediments with endogenous RioK3 on a sucrose gradient, data not shown) was subjected to ultracentrifugation in a sucrose gradient. The fractions containing pre-40S and 40S particles, checked for their content in HA-RioK3, were pooled and used in immunoprecipitation experiments performed with an anti-HA affinity matrix (). The co-precipitated RNAs were analyzed by real time RT-q-PCR () because the amount of RNA co-precipitated with the transiently expressed tagged-protein is too low to allow its analysis by Northern-blot. 18S-E pre-rRNA and 18S rRNA + 18S-E pre-rRNA were significantly amplified from IP samples obtained using the anti-HA matrix (BHA lanes) as compared with control samples from mock precipitation experiments omitting anti-HA antibodies (B0 lanes). In contrast, 28S rRNA and the Gapdh mRNA were not amplified from these samples (as expected since 40S fractions were used as starting material for these experiments). Comparison of the amplification levels obtained for 18S-E pre-rRNA and 18S rRNA + 18S-E pre-rRNA from the 40S Input fraction and from the HA-IP sample shows that the ratio 18S-E/(18S + 18S-E) is much more elevated in the IP sample: the ratio 18S-E/(18S + 18S-E) of ~0.25 observed in the Input increases to ~1.45 in the BHA fraction, corresponding to a more than 5x enrichment in 18S-E relative to 18S in the HA-RioK3 IP sample. These data strongly suggest that RioK3 is present in pre-40S particles containing the 18S-E pre-rRNA.
Figure 4. Immunoprecipitation of HA-RioK3. Fractions containing pre-40S and 40S particles, checked for their content in HA-RioK3 by western blot, were subjected to immunoprecipitation with an anti-HA affinity matrix. (A) western blot analysis of the efficiency of HA-RioK3 immunoprecipitation. Proteins were extracted from the agarose beads devoid (B0) or coated with anti-HA antibody (BHA) or from 1/2 of the input 40S fractions (Input) and subjected to western blot analysis. (B) Total RNAs extracted from the IP fractions (B0 and BHA) or from 1/5 of the input 40S fraction (Input) were reverse-transcribed as described in the Materials and Methods section, and 1/100 dilutions of the corresponding cDNAs were analyzed by real-time RT-q-PCR for the presence of 18S-E pre-rRNA, 18S and 28S rRNA and Gapdh mRNA. Quantities of each amplicon, expressed as arbitrary units, were obtained from standard curves determined for each RNA species as described in the Materials and Methods section. Values presented are the means of quadruplate experiments (+/−SEM). Pair-wise statistical analysis for (BHA) vs. (B0) was performed with Student’s t-test ***p < 0.001.
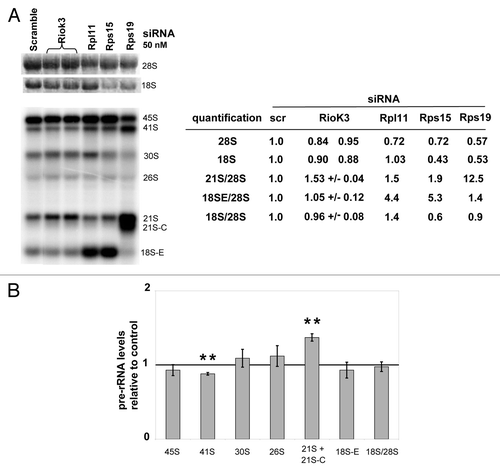
RioK3 depletion leads to a specific increase in 21S pre-rRNA levels
Requirement of RioK3 for ribosome synthesis was assessed by knocking-down expression of this protein with a siRNA treatment. RioK3 depletion 48 h after siRNA treatment is efficient (the protein level measured by western-Blot was decreased by ~90% as shown in ), with no clear modification in the ribosome profile () as compared with the profile obtained with an extract from scramble siRNA-treated cells (). We did not observe any modification in the amount of RioK2 () when RioK3 was depleted, while on the contrary, RioK2 depletion clearly affected RioK3 level () as well as the one of free 40S particles (). Since subtle effects of RioK3 depletion on pre-rRNA maturation cannot be excluded, we analyzed total cellular RNAs 48 h post-transfection with RioK3 siRNAs or either scramble, Rps15, Rps19 or Rpl11 siRNAs (). Mature 18S and 28S rRNA amounts were measured after agarose gel electrophoresis and ethidium bromide staining and amounts of 18S rRNA precursors were determined by northern blot analysis with a 5′ITS1 probe ( and Fig. S1 for the pre-rRNA processing scheme). The 18S/28S molar ratio was not found to change significantly when RioK3 was depleted, compared with Rps15, Rps19 or Rpl11 depletions. The most important effect we detected is the increased accumulation of the 21S intermediate (). Altogether, these results indicate that the absence of RioK3 leads to a subtle disturbance of the pre-rRNA processing pathway leading to 18S rRNA synthesis.
Figure 5. Impact of RioK3 depletion on ribosome biogenesis. (A) Analysis of rRNA and pre-rRNA levels in HeLa cells transfected with scramble or RioK3, Rpl11, Rps15, or Rps19 siRNAs. Identical amounts of RNAs (3 μg) from HeLa cells, extracted 48 h after transfection, were separated on a 1.2% agarose gel and then transferred to a Hybond N+ membrane. The mature rRNA species were revealed by ethidium bromide staining and the pre-rRNA species by hybridization with a 32P-labeled 5′-ITS1 probe. Mature rRNA, 21S and 18S-E pre-rRNA species were quantified. Pre-rRNA species are described in Fig. S1. (B) Levels of pre-rRNA intermediates in RioK3 siRNA-treated cells 48 h post transfection relative to those in cells treated with scramble siRNAs (arbitrarily set to 1). The means (+/− SEM) of 3 independent Northern experiments are presented. Pair-wise statistical analysis was performed with Student’s t-test: *p < 0.05; **p < 0.01; ***p < 0.001.
Discussion
The data presented here indicate that in HeLa cells, RioK3, the protein of the RIO kinase family found thus far only in multicellular eukaryotes, belongs to cytoplasmic pre-40S pre-ribosomal particles. It was found concentrated in the cytoplasm of HeLa cells and does not seem to shuttle between nucleus and cytoplasm via a Crm1-dependent mechanism, at least in the HeLa cell line used here. It sediments with 40S ribosomal particles in a sucrose gradient and when the small ribosomal subunit biogenesis is impaired by depletion of either Rps15, Rps19 or RioK2, we observed a decrease in the amount of RioK3, suggesting it may well be a component of a (pre)-40S ribosomal particle, as both mature and pre-40S pre-ribosomal particle levels are affected by these 3 depletions. The hypothesis that RioK3 belongs to pre-40S pre-ribosomal particles was supported by the co-immunoprecipitation of a fraction of RioK3 with hLtv1, hEnp1 () together with RioK2 which are non-ribosomal protein components of pre-40S pre-ribosomal particles containing the 18S-E pre-rRNA ( and refs. Citation35, Citation37). Moreover, we show that cytoplasmic 18S-E pre-rRNA is co-precipitated with transiently expressed HA-tagged RioK3 (). Altogether, these results indicate that RioK3 is a component of one or several cytoplasmic pre-40S particle(s) that contain 18S-E pre-rRNA and hLtv1, hEnp1, or RioK2, either together in the same particle and/or present only in a subset of RioK3-containing particles. Hence, human pre-40S particles transiently associate with at least two RIO family proteins, RioK2 and RioK3.
The role of RioK2 in human 40S subunit biogenesis has been investigated in depth.Citation38 It was demonstrated to be involved in a step that was described for the first time in human cells, the cytoplasmic maturation of the ultimate 18S rRNA precursor named 18S-E, which features a 20–30 nt extension downstream from the 3′ end of the 18S rRNA sequence (Fig. S1). In the absence of RioK2, the 18S-E intermediate accumulates in the cytoplasm, a phenotype that is similar to the accumulation of the 20S pre-rRNA observed in yeast cells depleted of the Rio2p protein. RioK2 was also recently demonstrated to be involved in the export of pre-40S particles as well as in the recycling into the nucleus of pre-40S particle components such as hEnp1/bystin, hDim2, hLtv1 and hNob1.Citation35 Until recently, no role in ribosome biogenesis was reported for the third human RIO family protein, RioK1. In our hands, depletion of RioK1 by RNA interference in HeLa cells did not lead to any obvious pre-rRNA maturation defect (unpublished data). However, RioK1 may play at least an indirect role in ribosome production since it was very recently proposed to be an adaptor recruiting nucleolin, an RNA-binding protein involved in ribosome synthesis, to the PRMT5 complex for its symmetrical arginine methylation.Citation48 Concerning RioK3, our results suggest that it only plays a subtle role in 40S subunit biogenesis, although we cannot formally exclude the possibility that the low levels of RioK3 remaining following depletion are sufficient to support some of its functions. Nevertheless, while RioK3 depletion did not affect the steady-state accumulation of mature 18S rRNA and 40S ribosomal subunits, it led to a specific increase in the levels of the 21S pre-rRNA, the levels of the other 18S rRNA precursors remaining unaffected. The accumulation of this 21S intermediate may be caused by a delay in the nucleoplasmic cleavage at site E in ITS1 (see Fig. S1). Since our results suggest that RioK3 is concentrated in the cytoplasm, the delayed processing of the 21S pre-rRNA may be the indirect consequence of reduced recycling of some cytoplasmic pre-40S particle components back into the nucleus, which for example occurs in cells depleted of RioK2 (see above). Nevertheless, we cannot totally exclude that RioK3 is present at very low level in the nucleus at steady-state and that its function in 21S processing is more direct.
When the biogenesis of the large ribosomal subunit is impaired (by depletion of Rpl11 or Rpl5), we observed a spectacular and specific increase in the steady-state levels of RioK3, which still exclusively sediments on sucrose gradients in the 40S fraction. Under these conditions, more RioK3 was co-immunoprecipitated together with hLtv1 (Fig. S2), suggesting that more pre-40S particles containing hLtv1 contain also RioK3. Moreover, 18S-E pre-rRNA was clearly and specifically co-precipitated (Fig. S2) Nevertheless, most of the RioK3 molecules present in the 40S fraction were not precipitated with anti-hLtv1 antibodies and may belong to late pre-40S particles in which hLtv1 (and hEnp1) are no longer present or RioK3 molecules are dissociated from the pre-40S particles during immunoprecipitation. The unexpected increase in RioK3 steady-state levels following Rpl11 or Rpl5 depletion may be due to a specific increase in the steady-state levels of RioK3 mRNA, and/or increased translation of this mRNA and/or stabilization of the protein. An increase in RioK3 mRNA levels is not the sole explanation, as in the absence of Rpl11, not only RioK3 but also RioK2 and hLtv1 mRNA levels were increased relative to Gapdh mRNA to the same moderate extent (Fig. S3). Another possibility is that RioK3 is rapidly turned over when not associated with cytoplasmic pre-40S particles; in such a scenario, its steady-state levels are expected to increase when those of cytoplasmic pre-40S particles rise, as is the case in cells depleted of Rpl11 which display a strong increase in 18S-E pre-rRNA amounts ().
Interestingly, it has recently been reported that the RIOK3 gene is overexpressed in pancreal ductal adenocarcinoma (PDAC), one of the most lethal cancers, and that knock-down of RIOK3 expression by shRNAs in PDAC cells reduces their ability to proliferate.Citation49,Citation50 It was also recently demonstrated that RIOK3 is upregulated in erythroblasts during terminal erythroid differentiation by downregulation of the microRNA miR-191Citation51 and that depletion of RIOK3 mRNAs led to impaired chromatin condensation and enucleation during terminal erythroid differentiation. It remains to be established whether these effects of RioK3 depletion on PDAC cell proliferation and terminal erythroid differentiation are due to impaired ribosome biogenesis and/or inhibition of another process in which RioK3 is implicated. While this work was under revision, Widmann and collaboratorsCitation52 published that RioK1 and RioK 3 are associated with pre-40S pre-ribosomal particles and that depletion of RioK1 affects the production of 18S rRNA and the recycling of several trans-acting factors.
Materials and Methods
Cell culture, plasmids and media
HeLa and U2OS cells were grown in Dulbecco Modified Eagle Medium (DMEM) supplemented with 10% fetal calf serum (Gibco), 1 mM sodium pyruvate, 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco) at 37°C with 5% CO2. LMB was purchased from LC Laboratories and used at 10 nM or 20 nM for 2 h. Plasmids pEGFP-RIOK3 and E46-HA-RIOK3 were constructed as follows: the RIOK3 coding sequence was amplified by PCR using an IMAGE clone containing the full length RIOK3 cDNA inserted in pCMVsport6-RIOK3 (clone BC039729 from Open Biosystems) and the primers K3(6)/K3(2) and K3(6)/K3(8), respectively (see Table S1). The DNA fragments amplified with the K3(6)/K3(2) and K3(6)/K3(8) primer pairs were respectively cloned in frame with the sequence encoding EGFP in plasmid pEGFP-C2 (Bioscience Clontech) using KpnI and BamHI restriction sites and with the sequence encoding the HA oligopeptide in plasmid E046 (a gift of D. Trouche) using KpnI and XbaI restriction sites. pEGFP-RI0K2 described in reference Citation38 was kindly provided by Pierre-Emmanuel Gleizes.
Transfection of HeLa cells, protein analysis by western blot
Exponentially growing adherent HeLa cells were transformed with plasmid DNAs by the “Calcium phosphate” methodCitation53 or with jetPRIME (Polyplus) according to the manufacturer’s recommendations. Transfected cells were grown for 24 or 48 h. Cells were collected following trypsin digestion (Invitrogen), washed 1 time in 5 mL Net2 buffer (50 mM Tris-HCl pH 7.4, 250 mM NaCl) containing a protease inhibitor cocktail (complete-EDTA free, Roche), pellets were resuspended in 400 μL Net2 buffer and subjected to sonication for 5 min (medium state, 30 sec ON, 30 sec Off; Bioruptor Diagenode). Extracts were recovered after a 10 min centrifugation at 13 000 rpm, 4°C. Protein concentrations were determined using the BioRad protein assay method. Samples corresponding to 40 μg of proteins were treated as described in the NuPAGE protocol (Invitrogen) and loaded on 8% pre-cast polyacrylamide-sodium dodecyl sulfate (SDS) gels (NuPAGE Novex Bis Tris midi gels, Invitrogen), and after migration, transferred to nitrocellulose membranes using an iBlot dry blotting system (Invitrogen). RioK3 and RioK2 were detected with mouse monoclonal anti-RioK3 (H00008780-MO1, Abnova) and mouse polyclonal anti-RioK2 antibodies (H00055781-B01P Abnova) diluted at 1:1000 and 1:500 respectively in PBST buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, 0.1% Tween 20) containing 5% (w/v) milk. HA oligopeptide was detected with a 1/1000 dilution of a horseradish-peroxidase (HRP)-conjugated mouse monoclonal anti-HA-antibody (clone 3F10, Roche). Actin was detected with a 1/10000 dilution of a mouse monoclonal anti-actin antibody (Millipore). hLtv1 and hEnp1 were respectively detected with rabbit polyclonal anti-hLtv1 (diluted at 1:5000) and anti-hEnp1 (diluted at 1:1000) antibobies kindly provided by Marlène Faubladier (LBME-CNRS, Toulouse). Rps19 was detected with a rabbit polyclonal anti-Rps19 antibody diluted at 1:1000. For protein detection, membranes were saturated by a 1 h incubation with PBST buffer containing 5% milk followed by a 1 h-15 h incubation with the primary antibodies, followed by three washing steps in PBST buffer, and a 1 h incubation with the relevant HRP-conjugated secondary antibodies (anti-mouse or anti-rabbit, Promega) diluted to 1:10000 in PBST buffer containing 5% milk, and finally washed three times for 10 min with PBST buffer. Detections were achieved by using either regular enhanced chemoluminescence (ECL) western blotting detection reagents (GE Healthcare) or a more sensitive Lumilight plus (Roche) western blotting detection kit. Chemoluminescence signals were analyzed with a BioImager (FujiFilm, LAS 4000) and quantified with the MultiGauge software.
Immunofluorescence microscopy
HeLa cells growing exponentially on microscope cover glasses in 6-well plates were transformed with pEGFP-RIOK3 or pEGFP-RIOK2 by the Ca-Phosphate method. After 48 h, cover glasses were washed twice in PBS and cells were fixed for 30 min with 4% paraformaldehyde in PBS. Fixed cells were washed twice with PBS. DNA was counterstained with Hoechst 33342 (Molecular Probes) for 10 min at room temperature. The coverslips were then rinsed twice in PBS and mounted in Mowiol 4.88 (Polyscience Inc., Eppelheim, Germany). Images were captured by laser-scanning confocal microscopy (Leica SP2) using the Metavue software.
Knockdown of gene expression by RNA interference (RNAi)
The siRNA duplexes (Table S1) and siRNA-negative control duplex (scramble siRNA) were purchased from Eurogentec. Transfection of HeLa cells with siRNAs (10 to 50 nM) was performed using Interferin (PolyPlus) according to the manufacturer’s recommendations. Control samples were transfected with a scramble siRNA (siRNA-negative control duplex; Eurogentec). The efficiency of the downregulation induced by each siRNA was assessed by a quantitative western blot analysis taking actin as an internal control to normalize expression of the target genes.
Cell fractionation
Cells were washed successively with DMEM, PBS and incubated 10 min on ice in buffer A (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl). Mechanical disruption was performed with a Dounce homogenizer in Buffer A containing 0.5 mM DTT. After centrifugation at 1000 g for 10 min at 4°C, the supernatant, considered the cytoplasmic fraction, was collected and the pellet, containing the nuclei, was washed with 10 mM Tris, pH 7.5, 33 mM MgCl2, 250 mM sucrose. The nuclear fraction was resuspended in 10 mM MgCl2, 250 mM sucrose and further purified by centrifugation for 10 min at 500 g on a sucrose cushion (0.5 mM MgCl2, 350 mM sucrose). The nuclei were then lysed in 50 mM Na acetate, pH 5.1, 140 mM NaCl, 0.3% SDS. Aliquots of each fraction were analyzed by western blot after SDS-PAGE electrophoresis (NuPAGE Novex Bis Tris midi gels, Invitrogen) using anti-RioK3 antibody as described above and rabbit polyclonal anti-dyskerin antibodyCitation54 at 1:1000 dilution.
Total RNA extractions and northern hybridizations
Total RNA extractions were performed using Trizol (Invitrogen) according to the manufacturer’s protocol. Total RNAs (~4 μg/well) or half of the RNA samples purified from 250 μL of the sucrose gradient fractions were mixed with 3 volumes of loading dye (DMSO 60% (v/v), deionised Glyoxal (Sigma) 20% (v/v), Glycerol 5% (v/v), ethidium bromide 0.2 mg/mL in BPTE buffer (PIPES 10 mM, Bis-TRIS 30 mM, EDTA 1 mM pH8), and incubated 1 h at 55°C. Electrophoresis of glyoxylated RNAs in 1.2% agarose gels was performed as reported (Sambrook and Russel, 2001), and RNAs were transferred to Amersham Hybond N+ membranes (GE Healthcare). Mature rRNAs were analyzed by ethidium bromide staining using a Gel-doc (BioRad), and pre-rRNA precursors by Northern hybridization of the membranes with a 32P-labeled ITS1 oligodeoxynucleotide probe (Table S1) using the Rapid Hyb Buffer protocol (GE Healthcare).
Cell extract preparation and sucrose gradient sedimentation experiments
Cells growing exponentially (1 × 108 cells per gradient) were treated for 10 min with 50 μg/mL cycloheximide (Sigma) directly added in the culture medium, then collected following trypsin digestion, washed with PBS buffer supplemented with cycloheximide, and harvested by centrifugation at 500 g for 5 min at 4°C. The dry pellet was resuspended in 1 volume of lysis buffer (50 mM Tris-HCl, pH 7.4, 50 mM KCl, 10 mM MgAc, 50 mg/mL cycloheximide) supplemented with 1 × complete EDTA-free protease inhibitor cocktail (Roche), 1 mM dithiothreitol and 0.1 unit/μL RNasin (Promega). About 200 μL of ice-cold glass beads were added to 1 mL of cell suspension. Cells were broken by vigorous vortexing at 4°C, (3 × 1 min separated by 1 min pauses). Lysis of cells was checked by optical microscopy. After a first centrifugation for 10 min at 3000 rpm to eliminate the glass beads, the supernatant or extract was clarified by centrifugation for 10 min at 13000 rpm at 4°C. Protein concentrations were determined using the BioRad protein assay method. The equivalent of 2 mg of proteins in 500 μL of sample was layered on a 15–50% (w/v) sucrose gradient prepared in sucrose buffer (50 mM TRIS-acetate, pH 7.5, 50 mM NH4Cl, 12 mM Mg acetate, 1 mM dithiothreitol) with a Gradient Master Former (Biocomp Instruments) and centrifuged in polyallomer centrifuge tubes (14 × 89 mm, Beckman) for 15 h at 33000 rpm and 4°C in an Optima L-100XP ultracentrifuge (Beckman Coulter) using the SW41 rotor. Following centrifugation, 18 fractions of 500 μL each were collected from the top of the gradient with a Foxy Jr gradient fraction collector (Teledyne ISCO). Absorbance at 254 nm was recorded during collection with a UA-6 device (Teledyne ISCO). For protein analysis by western blot, 250 µL of each fraction were precipitated with trichloroacetic acid (final concentration 15% v/v), washed with cold acetone, dried at room temperature and resuspended with 30 μL loading buffer. Protein samples were treated according to the NuPAGE protocol (Invitrogen), loaded on SDS-PAGE and analyzed by western blot as described above. For RNA analysis, 150 μL of gradient fractions were extracted using GTC mix (250 μL of 4 M guanidinium isothiocyanate mix in 10 mM TRIS-HCl (pH 8.0), 1 mM EDTA), 2 μL of glycogen (20 mg/mL), 150 μL of 100 mM NaAc (pH 5), 225 μL of phenol and 225 μL of chloroform. The samples were thoroughly mixed, incubated 5 min at 65°C, centrifuged 5 min at 16000 g at 4°C. 350 μL of the aqueous phase were then precipitated with 2 vol ethanol, centrifuged, dried, dissolved in 10 μL nuclease-free water and analyzed by northern blot.
Preparative 15–50% (w/v) sucrose gradients were poured/freezed in 4 successive layers (50%, 40%, 30%, 15% sucrose, 9 mL each) in polyallomer centrifuge tubes (25 × 89 mm, 38 mL, Beckman). An amount of commercial Hela cytoplasmic extract (CILbiotech; B-7000 Mons) corresponding to 25 mg of proteins was loaded on a defrosted sucrose gradient and tubes were centrifuged at 4°C at 32000 rpm for 16 h in a SW32 rotor in an Optima L-100XP ultracentrifuge (Beckman Coulter).
Immunoprecipitations and RT-q-PCR experiments
Immunoprecipitations of hLtv1 and hEnp1 were performed using aliquots of 40S fractions prepared from commercial HeLa cytoplasmic extracts. Anti-hLtv1 and hEnp1 coated beads were prepared as follows: for each IP, 50 μL of proteinA-Sepharose beads (GE Healthcare) equilibrated in sucrose buffer (50 mM Tris-acetate, pH 7.5, 50 mM NH4Cl, 12 mM Mg acetate, 1 mM dithiothreitol) supplemented with 1 × complete EDTA-free protease inhibitor cocktail (Roche) and 0.1 U/ μL RNasin (Promega) were incubated with 20 μL of anti-hLtv1, anti-hEnp1 or non-immune serum in a total volume of 500 μL of sucrose buffer, during 1 h at 4°C on a rotating wheel. After a 2 min centrifugation (10000 rpm) to eliminate the flow-through, followed by 4 washes of the beads using 1 mL of ice-cold sucrose buffer, the beads were incubated with 400 μL of pooled 40S fractions in a total volume of 1.2 mL sucrose buffer, at 4°C during 1 h on a rotating wheel. After elimination of the flow through, bead pellets were washed 5 times with 1 mL of ice-cold sucrose buffer, and during the last washing step, bead suspensions were divided into two halves and treated for protein and RNA analysis as described for the HA-RioK3 IP.
To immunoprecipitate HA-RioK3, a cytoplasmic extract prepared from transiently transfected cells as described above corresponding to about 4 mg of protein was subjected to ultracentrifugation in sucrose gradient, and the fractions containing pre-40S and 40S particles, checked for their content in HA-RioK3, were pooled. Aliquots corresponding to 1/3 of the 40S pool were incubated by rotation on a wheel for 1 h at 4°C with 100 μL (bead volume) of either control agarose resin (Pierce) (B0) or anti-HA affinity Matrix (Roche) (BHA) in a total volume of 1.5 mL. Beads were pre-equilibrated and the incubation volume adjusted with IP 200 buffer (20 mM TRIS-HCl pH 7.5, 200 mM NaCl, 5 mM Mg acetate, 1 mM dithiothreitol, 0.1% Triton X100) supplemented with 1 × complete EDTA-free protease inhibitor cocktail (Roche) and [0.1 U/ μL RNasin (Promega)]. Flow-through were collected after 1 min 30 sec centrifugation at 10 000 rpm, and the bead pellets were washed 6 times with 1 mL of ice-cold IP 200 buffer. During the last washing, bead suspensions were divided into two aliquots: a 200 µl aliquot for protein analysis and a 800 µl aliquot transferred to a new tube for RNA analysis. After a last centrifugation step, the remaining buffer was carefully discarded from the aliquot for protein analysis and the 20 μL dried beads were directly resuspended in 30 μL of NuPAGE lithium dodecyl sulfate sample buffer supplemented with NuPAGE sample-reducing agent (Invitrogen), heated 10 min at 70°C, and 15 µl loaded on NuPAGE Novex 8% Bis-Tris gels (Invitrogen), together with an aliquot corresponding to ¼ of the Input analyzed after treatment by the beads, and transferred to nitrocellulose membranes for western blot analysis. The aliquot for RNA analysis was treated as follows. Bead pellets (80 μL) were resuspended in 150 μL of IP 200 buffer and extracted using the GTC mix as described above for gradient fractions. RNA pellets were resuspended in 40 μL H2O and treated with 1 μL DNase (Promega) for 1 h at 37°C. Reaction was stopped with 5 μL of stop DNase buffer and 50 μL of saturated phenol (pH 4.5). The mixture was vigorously shaken and centrifuged 5 min at 13000 rpm and at 4°C. 45 μL of aqueous phase were kept on ice. The phenolic phase was washed using 50 μL H2O and this second aqueous phase was pooled with the first one. This combined aqueous phase was extracted using 90 μL of chloroform and 85 μL of aqueous phase were precipitated with 500 μL ethanol, 9 μL 3 M sodium acetate and 2 μL glycogen at 20 mg/mL. After 5 min centrifugation at 13000 rpm, 4°C, the RNA pellets were air-dried and resuspended in 10 μL of nuclease-free water. An aliquot of the input 40S fraction corresponding to 1/5 of the amount immunoprecipitated on beads was DNase-treated in parallel. Half of the aliquots of the DNase-treated RNAs were reverse transcribed using the Superscript II reverse transcriptase (Invitrogen) and random primers following the protocol provided by the enzyme’s manufacturer. Following cDNA synthesis, RNAs were hydrolyzed for 15 min at 70°C in the presence of 115 mM NaOH. Samples were neutralized with 115 mM HCl and the cDNAs were analyzed by RT-q-PCR on a Mastercycler ep realplex (Eppendorf). Each RT-q-PCR reaction was performed in triplicate or quadruplate in 15 µl final volume containing 7.5 µl of the iQ SYBR Green Supermix (Bio-Rad), 0.5 µM of each specific oligo and 5 µl of a 1/100 dilution of the cDNA samples obtained from reverse transcription of the Input, B0 and BHA RNA aliquots. The cycle parameters were: 10 min at 95°C as pre-amplification step, 15 sec at 95°C–15 sec at 55°C–30 sec at 68°C for 40 cycles as amplification steps, and warming from 60°C to 95°C for the melting curve. Specificity for each RT-PCR reaction was confirmed by the observation of only one melting peak between 80–90°C depending on the length of the amplified fragment. The levels of each cDNA, expressed in arbitrary units, were calculated from standard curves fitted to a logarithmic function, obtained from 6 different appropriate serial dilutions of the cDNAs synthesized from total RNAs purified from the starting HeLa cytoplasmic extract. Efficiencies calculated from these curves ranged between 53 and 72%. The primers used for RT-q-PCR are indicated in Table S1.
Additional material
Download Zip (2.4 MB)Acknowledgments
We are very grateful to Anaïs Vaissière for help with some immunoprecipitation and RT-PCR experiments and Didier Trouche for gift of plasmid E046. We acknowledge Pierre Emmanuel Gleizes, Coralie Carron, Valérie Cadamuro, Marie-Françoise O’Donohue and Chrystelle Bonnart for reagents and help with microscopy and cell culture. We thank all members of the group of M.C.-F. and Y.H., especially Jean-Paul Gélugne for advice and helpful discussion. We thank Jean-Paul Gélugne and Anthony Henras for critical reading of the manuscript, David Vila for help with figures, and Ulrike Kutay, Barbara Widmann and Yvo Zemp for discussion and sharing unpublished results.
Financial support
This work was supported by the CNRS, Université Paul Sabatier, and grants from the Agence Nationale de la Recherche and the Ligue Nationale Contre le Cancer (“équipe labellisée”).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Warner JR. Nascent ribosomes. Cell 2001; 107:133 - 6; http://dx.doi.org/10.1016/S0092-8674(01)00531-1; PMID: 11672521
- Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene 2003; 313:17 - 42; http://dx.doi.org/10.1016/S0378-1119(03)00629-2; PMID: 12957375
- Schäfer T, Strauss D, Petfalski E, Tollervey D, Hurt E. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J 2003; 22:1370 - 80; http://dx.doi.org/10.1093/emboj/cdg121; PMID: 12628929
- Tschochner H, Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol 2003; 13:255 - 63; http://dx.doi.org/10.1016/S0962-8924(03)00054-0; PMID: 12742169
- Milkereit P, Kühn H, Gas N, Tschochner H. The pre-ribosomal network. Nucleic Acids Res 2003; 31:799 - 804; http://dx.doi.org/10.1093/nar/gkg165; PMID: 12560474
- Panse VG, Johnson AW. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem Sci 2010; 35:260 - 6; http://dx.doi.org/10.1016/j.tibs.2010.01.001; PMID: 20137954
- Granneman S, Baserga SJ. Crosstalk in gene expression: coupling and co-regulation of rDNA transcription, pre-ribosome assembly and pre-rRNA processing. Curr Opin Cell Biol 2005; 17:281 - 6; http://dx.doi.org/10.1016/j.ceb.2005.04.001; PMID: 15901498
- Zemp I, Kutay U. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett 2007; 581:2783 - 93; http://dx.doi.org/10.1016/j.febslet.2007.05.013; PMID: 17509569
- Li Z, Lee I, Moradi E, Hung NJ, Johnson AW, Marcotte EM. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol 2009; 7:e1000213; http://dx.doi.org/10.1371/journal.pbio.1000213; PMID: 19806183
- Henras AK, Soudet J, Gérus M, Lebaron S, Caizergues-Ferrer M, Mougin A, et al. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 2008; 65:2334 - 59; http://dx.doi.org/10.1007/s00018-008-8027-0; PMID: 18408888
- Strunk BS, Karbstein K. Powering through ribosome assembly. RNA 2009; 15:2083 - 104; http://dx.doi.org/10.1261/rna.1792109; PMID: 19850913
- Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol Cell 2002; 10:105 - 15; http://dx.doi.org/10.1016/S1097-2765(02)00579-8; PMID: 12150911
- Ho Y, Mason S, Kobayashi R, Hoekstra M, Andrews B. Role of the casein kinase I isoform, Hrr25, and the cell cycle-regulatory transcription factor, SBF, in the transcriptional response to DNA damage in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 1997; 94:581 - 6; http://dx.doi.org/10.1073/pnas.94.2.581; PMID: 9012827
- Vanrobays E, Gleizes PE, Bousquet-Antonelli C, Noaillac-Depeyre J, Caizergues-Ferrer M, Gélugne JP. Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J 2001; 20:4204 - 13; http://dx.doi.org/10.1093/emboj/20.15.4204; PMID: 11483523
- Vanrobays E, Gelugne JP, Gleizes PE, Caizergues-Ferrer M. Late cytoplasmic maturation of the small ribosomal subunit requires RIO proteins in Saccharomyces cerevisiae. Mol Cell Biol 2003; 23:2083 - 95; http://dx.doi.org/10.1128/MCB.23.6.2083-2095.2003; PMID: 12612080
- Lafontaine D, Vandenhaute J, Tollervey D. The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev 1995; 9:2470 - 81; http://dx.doi.org/10.1101/gad.9.20.2470; PMID: 7590228
- Lafontaine DL, Preiss T, Tollervey D. Yeast 18S rRNA dimethylase Dim1p: a quality control mechanism in ribosome synthesis?. Mol Cell Biol 1998; 18:2360 - 70; PMID: 9528805
- Fatica A, Oeffinger M, Dlakicá M, Tollervey D. Nob1p is required for cleavage of the 3′ end of 18S rRNA. Mol Cell Biol 2003; 23:1798 - 807; http://dx.doi.org/10.1128/MCB.23.5.1798-1807.2003; PMID: 12588997
- Fatica A, Tollervey D, Dlakicá M. PIN domain of Nob1p is required for D-site cleavage in 20S pre-rRNA. RNA 2004; 10:1698 - 701; http://dx.doi.org/10.1261/rna.7123504; PMID: 15388878
- Pertschy B, Schneider C, Gnädig M, Schäfer T, Tollervey D, Hurt E. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J Biol Chem 2009; 284:35079 - 91; http://dx.doi.org/10.1074/jbc.M109.040774; PMID: 19801658
- Vanrobays E, Gélugne JP, Caizergues-Ferrer M, Lafontaine DL. Dim2p, a KH-domain protein required for small ribosomal subunit synthesis. RNA 2004; 10:645 - 56; http://dx.doi.org/10.1261/rna.5162204; PMID: 15037774
- Vanrobays E, Leplus A, Osheim YN, Beyer AL, Wacheul L, Lafontaine DL. TOR regulates the subcellular distribution of DIM2, a KH domain protein required for cotranscriptional ribosome assembly and pre-40S ribosome export. RNA 2008; 14:2061 - 73; http://dx.doi.org/10.1261/rna.1176708; PMID: 18755838
- Oeffinger M, Dlakic M, Tollervey D. A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes Dev 2004; 18:196 - 209; http://dx.doi.org/10.1101/gad.285604; PMID: 14729571
- Chen W, Bucaria J, Band DA, Sutton A, Sternglanz R. Enp1, a yeast protein associated with U3 and U14 snoRNAs, is required for pre-rRNA processing and 40S subunit synthesis. Nucleic Acids Res 2003; 31:690 - 9; http://dx.doi.org/10.1093/nar/gkg145; PMID: 12527778
- Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 2002; 417:967 - 70; http://dx.doi.org/10.1038/nature00769; PMID: 12068309
- Loar JW, Seiser RM, Sundberg AE, Sagerson HJ, Ilias N, Zobel-Thropp P, et al. Genetic and biochemical interactions among Yar1, Ltv1 and Rps3 define novel links between environmental stress and ribosome biogenesis in Saccharomyces cerevisiae. Genetics 2004; 168:1877 - 89; http://dx.doi.org/10.1534/genetics.104.032656; PMID: 15611164
- Gelperin D, Horton L, Beckman J, Hensold J, Lemmon SK. Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA 2001; 7:1268 - 83; http://dx.doi.org/10.1017/S1355838201013073; PMID: 11565749
- Granneman S, Nandineni MR, Baserga SJ. The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14. Mol Cell Biol 2005; 25:10352 - 64; http://dx.doi.org/10.1128/MCB.25.23.10352-10364.2005; PMID: 16287850
- Lebaron S, Papin C, Capeyrou R, Chen YL, Froment C, Monsarrat B, et al. The ATPase and helicase activities of Prp43p are stimulated by the G-patch protein Pfa1p during yeast ribosome biogenesis. EMBO J 2009; 28:3808 - 19; http://dx.doi.org/10.1038/emboj.2009.335; PMID: 19927118
- Schäfer T, Maco B, Petfalski E, Tollervey D, Böttcher B, Aebi U, et al. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature 2006; 441:651 - 5; http://dx.doi.org/10.1038/nature04840; PMID: 16738661
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, et al. Global analysis of protein localization in budding yeast. Nature 2003; 425:686 - 91; http://dx.doi.org/10.1038/nature02026; PMID: 14562095
- Seiser RM, Sundberg AE, Wollam BJ, Zobel-Thropp P, Baldwin K, Spector MD, et al. Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae. Genetics 2006; 174:679 - 91; http://dx.doi.org/10.1534/genetics.106.062117; PMID: 16888326
- Lamanna AC, Karbstein K. Nob1 binds the single-stranded cleavage site D at the 3′-end of 18S rRNA with its PIN domain. Proc Natl Acad Sci U S A 2009; 106:14259 - 64; http://dx.doi.org/10.1073/pnas.0905403106; PMID: 19706509
- Lamanna AC, Karbstein K. An RNA conformational switch regulates pre-18S rRNA cleavage. J Mol Biol 2011; 405:3 - 17; http://dx.doi.org/10.1016/j.jmb.2010.09.064; PMID: 20934433
- Zemp I, Wild T, O’Donohue MF, Wandrey F, Widmann B, Gleizes PE, et al. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J Cell Biol 2009; 185:1167 - 80; http://dx.doi.org/10.1083/jcb.200904048; PMID: 19564402
- Léger-Silvestre I, Caffrey JM, Dawaliby R, Alvarez-Arias DA, Gas N, Bertolone SJ, et al. Specific Role for Yeast Homologs of the Diamond Blackfan Anemia-associated Rps19 Protein in Ribosome Synthesis. J Biol Chem 2005; 280:38177 - 85; http://dx.doi.org/10.1074/jbc.M506916200; PMID: 16159874
- Carron C, O'Donohue MF, Choesmel V, Faubladier M, Gleizes PE. Analysis of two human pre-ribosomal factors, bystin and hTsr1, highlights differences in evolution of ribosome biogenesis between yeast and mammals. Nucleic Acids Res 2011; 39:280 - 91; http://dx.doi.org/10.1093/nar/gkq734; PMID: 20805244
- Rouquette J, Choesmel V, Gleizes PE. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J 2005; 24:2862 - 72; http://dx.doi.org/10.1038/sj.emboj.7600752; PMID: 16037817
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science 2002; 298:1912 - 34; http://dx.doi.org/10.1126/science.1075762; PMID: 12471243
- Angermayr M, Roidl A, Bandlow W. Yeast Rio1p is the founding member of a novel subfamily of protein serine kinases involved in the control of cell cycle progression. Mol Microbiol 2002; 44:309 - 24; http://dx.doi.org/10.1046/j.1365-2958.2002.02881.x; PMID: 11972772
- Johnson LN, Lowe ED, Noble ME, Owen DJ. The Eleventh Datta Lecture. The structural basis for substrate recognition and control by protein kinases. FEBS Lett 1998; 430:1 - 11; http://dx.doi.org/10.1016/S0014-5793(98)00606-1; PMID: 9678585
- Johnson LN, Noble ME, Owen DJ. Active and inactive protein kinases: structural basis for regulation. Cell 1996; 85:149 - 58; http://dx.doi.org/10.1016/S0092-8674(00)81092-2; PMID: 8612268
- LaRonde-LeBlanc N, Wlodawer A. The RIO kinases: an atypical protein kinase family required for ribosome biogenesis and cell cycle progression. Biochim Biophys Acta 2005; 1754:14 - 24; PMID: 16182620
- Krupa A, Srinivasan N. Lipopolysaccharide phosphorylating enzymes encoded in the genomes of Gram-negative bacteria are related to the eukaryotic protein kinases. Protein Sci 2002; 11:1580 - 4; http://dx.doi.org/10.1110/ps.3560102; PMID: 12021457
- Leonard CJ, Aravind L, Koonin EV. Novel families of putative protein kinases in bacteria and archaea: evolution of the “eukaryotic” protein kinase superfamily. Genome Res 1998; 8:1038 - 47; PMID: 9799791
- LaRonde-LeBlanc N, Wlodawer A. A family portrait of the RIO kinases. J Biol Chem 2005; 280:37297 - 300; http://dx.doi.org/10.1074/jbc.R500013200; PMID: 16183636
- Thomas F, Kutay U. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J Cell Sci 2003; 116:2409 - 19; http://dx.doi.org/10.1242/jcs.00464; PMID: 12724356
- Guderian G, Peter C, Wiesner J, Sickmann A, Schulze-Osthoff K, Fischer U, et al. RioK1, a new interactor of protein arginine methyltransferase 5 (PRMT5), competes with pICln for binding and modulates PRMT5 complex composition and substrate specificity. J Biol Chem 2011; 286:1976 - 86; http://dx.doi.org/10.1074/jbc.M110.148486; PMID: 21081503
- Kimmelman AC, Hezel AF, Aguirre AJ, Zheng H, Paik JH, Ying H, et al. Genomic alterations link Rho family of GTPases to the highly invasive phenotype of pancreas cancer. Proc Natl Acad Sci U S A 2008; 105:19372 - 7; http://dx.doi.org/10.1073/pnas.0809966105; PMID: 19050074
- Kalinina T, Güngör C, Thieltges S, Möller-Krull M, Penas EM, Wicklein D, et al. Establishment and characterization of a new human pancreatic adenocarcinoma cell line with high metastatic potential to the lung. BMC Cancer 2010; 10:295; http://dx.doi.org/10.1186/1471-2407-10-295; PMID: 20553613
- Zhang L, Flygare J, Wong P, Lim B, Lodish HF. miR-191 regulates mouse erythroblast enucleation by down-regulating Riok3 and Mxi1. Genes Dev 2011; 25:119 - 24; http://dx.doi.org/10.1101/gad.1998711; PMID: 21196494
- Widmann B, Wandrey F, Badertscher L, Wyler E, Pfannstiel J, Zemp I, Kutay U. The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40S subunits. Mol Biol Cell 2012; 23:22 - 35; PMID: 22072790
- Graham FL, van der Eb AJ. Transformation of rat cells by DNA of human adenovirus 5. Virology 1973; 54:536 - 9; http://dx.doi.org/10.1016/0042-6822(73)90163-3; PMID: 4737663
- Hoareau-Aveilla C, Bonoli M, Caizergues-Ferrer M, Henry Y. hNaf1 is required for accumulation of human box H/ACA snoRNPs, scaRNPs, and telomerase. RNA 2006; 12:832 - 40; http://dx.doi.org/10.1261/rna.2344106; PMID: 16601202
- LaRonde-LeBlanc N, Wlodawer A. Crystal structure of A. fulgidus Rio2 defines a new family of serine protein kinases. Structure 2004; 12:1585 - 94; http://dx.doi.org/10.1016/j.str.2004.06.016; PMID: 15341724
- LaRonde-LeBlanc N, Guszczynski T, Copeland T, Wlodawer A. Autophosphorylation of Archaeoglobus fulgidus Rio2 and crystal structures of its nucleotide-metal ion complexes. FEBS J 2005; 272:2800 - 10; http://dx.doi.org/10.1111/j.1742-4658.2005.04702.x; PMID: 15943813