Abstract
DNA sequences associated with protein-coding genes have been the primary focus of most genetic analyses of complex human diseases. Although we are rapidly gaining a comprehensive view of the etiology of certain central nervous system disorders, major gaps in our understanding persist. Recent studies have uncovered that many human genomic sequences are transcribed but not translated, generating an astounding diversity of noncoding RNAs (ncRNAs). This awareness should be taken into account when studying human diseases and may have profound implications on the development of novel biomarkers as well as therapies.
Introduction
The majority of transcription in humans results in the production of long non-protein-coding RNAs (lncRNAs), ranging from 200 bp up to several kilobases long. LncRNAs have only recently emerged as a major category of regulatory eukaryotic transcripts and can be classified into groups based on their location within genome. Natural antisense transcripts (NATs) arise from the opposite strand of protein-coding or non protein-coding genesCitation1-Citation7 and can overlap with exons, promoter or other regulatory elements of their cognate genes. Another class originates from the introns, promoters or 3′ termini of protein-coding genes.Citation8-Citation10 Finally, long intergenic RNAs (lincRNAs) are transcripts originating from genomic regions outside the boundaries of protein-coding genes.Citation11 Next generation sequencing of the human transcriptome has unearthed tens of thousands of lncRNAs from each class that are transcribed from specific cell and tissue types. As of February 2012, approximately 200 lncRNAs have been functionally annotated in the long noncoding RNA database (www.lncrnadb.com).Citation12 Although relatively few lncRNAs have been functionally characterized, increasing evidence suggests an important role for the thousands of uncharacterized remaining transcripts.
LncRNAs can have diverse functions including the ability to modulate transcription, regulate post-transcriptional RNA processing,Citation13 translation,Citation14 DNA methylation and chromatin architecture through local (cis) and long distance (trans) mechanisms.Citation15 Through interactions with transcription factors, co-activators and/or repressors, lncRNAs can affect different aspects of gene transcription to form a fine-tuned complex regulatory network. LncRNAs can modulate the activity of transcriptional machinery in cis in two main ways: by directly affecting transcription factors and their association with co-regulatorsCitation16-Citation18 or by orchestrating chromatin-remodeling events.Citation19-Citation21 Perhaps the most prominent mechanism involving lncRNAs is the recruitment of chromatin remodeling complexes, e.g., histone methyltransferases that methylate histones to either activate or repress gene transcription, in specific genomic loci. Interestingly, chromatin-remodeling proteins do not possess DNA-binding activity, suggesting that specific ncRNAs may be the critical link between these enzymes and the genome. In contrast to cis-acting lncRNAs, some transcripts exert their function by modulating gene expression on genomic loci other than the one from which they originate (trans activity).Citation11,Citation22,Citation23
Altered Expression of lncRNAs in CNS Diseases
Central nervous system (CNS) development, synaptic plasticity and the stress response are all tightly linked to epigenetic mechanisms that drive gene expression.Citation24 Temporal and spatial expression of lncRNAs appears to be important for proper neurological functioning through the precise regulation of epigenetic events. Many lncRNAs are expressed in the CNS where they have been shown to play fundamental roles.Citation15,Citation25
MALAT-1 is a lncRNA that was originally described as being overexpressed in different metastatic tumors, however it has recently been found to play a role in synapse formation.Citation26MALAT-1 is expressed in a variety of tissues but is enriched in the brain where it controls the expression of a subset of genes significantly associated with synaptogenesis. One of the hallmarks of human brain evolution is the expanding complexity of synaptic networks and the controlled expression of specific lncRNAs in neuronal cells.Citation27,Citation28 Transcriptomics studies have revealed the dynamic expression profile of several lncRNAs during the maturation of neuronal cell subtypes.Citation29 Some studies revealed the presence of “brain-associated” lncRNAs that affect hippocampal development, oligodendrocyte myelination and aging in the brain.Citation30 Thus, it is not surprising that the alteration of lncRNAs affects the basic functions of neuronal cells resulting in neurological and psychiatric disorders.Citation31-Citation33
In addition, hundreds of small non-coding RNA molecules have been identified in the mammalian central nervous system. Among these are microRNAs (miRNAs) which have demonstrated importance in many aspects of neuronal function. Disruption of gene regulation mediated by miRNA has been implicated in a range of disorders such as neurodegenerative and neuropsychiatric conditions as well as in tumors of the CNS. The focus of this review is to highlight the possible roles of the less explored long-noncoding RNAs in diseases of the CNS. For a comprehensive review of microRNA and other small RNA implicated in brain development and dysfunctions we refer the reader to other publications.Citation34-Citation37
LncRNAs in microsatellite expansion diseases
Microsatellite expansion (or trinucleotide repeats) give rise to more than 30 neurological and neuromuscular diseases, including: Huntington disease (HD), Fragile X syndrome (FXS) and Spinocerebellar Ataxias.Citation38,Citation39 Transcriptomics efforts have revealed that antisense transcription is a common feature of mammalian genes that are actively transcribed from microsatellite disease loci.Citation9,Citation40,Citation41 Transcription of lncRNAs containing repeats might alter their function and interfere with their ability to recruit protein complexes. The presence of nucleotide repeats might also alter the basal expression of lncRNAs, leading to an imbalance in sense-antisense transcript levels. Here, we highlight some of the recent findings showing that altered expression of specific lncRNAs might be involved in the etiology of several neurological disorders.
Fragile X syndrome
Fragile X syndrome (FXS) is the most common human heritable mental disorder. Patients display autistic symptoms and often suffer from epilepsy. A triplet nucleotide repeat expansion (> 200 repeats) in the 5′UTR of FMR1, the gene that encodes the crucial neuronal development protein, FMRP, is the causal factor in FXS (). This repeat expansion results in the methylation of the promoter and subsequent silencing of the FMR1 gene. Recent studies have suggested that noncoding RNAs may play a role in FXS as several lncRNAs are generated from the FMR1 gene locus.Citation9,Citation10 The long antisense RNA originating from intron 2, ASFMR1, has multiple alternative splicing patterns and spans the long repeat sequence ending in the gene promoter.Citation9 Another natural antisense transcript, FMR4, (~2 kb long, unspliced transcript) is initiated upstream of the FMR1 start site.Citation10 Previously, we have described that FMR4 plays a role in cell survival as FMR4 overexpression increases cell proliferation while FMR4 downregulation induces apoptosis.Citation10 We did not observe a cis acting effect on the expression of the FMR1 gene and thus hypothesized that the ability of FMR4 to interfere with proliferation pathways might occur in trans, however this has not yet been clarified. Moreover, FMR4, like FMR1, is silenced in Fragile X patients, thus its absence in the neurons of affected patients might contribute to the pathogenesis of this neurological disorder.
Figure 1. Microsatellite susceptible genes are featured by bidirectional transcription. (A-B) Fragile X Syndrome. (A) In general population the gene FMR1 contains at the 5′UTR up to 54 CGG repeat that do not interfere with the expression of the gene. In this locus, in people not affected by Fragile X syndrome, antisense noncoding RNAs have been reported to be transcribed from intron 2 (AS-FMR1) and from the promoter (FMR4). (B) Amplification of CGG trinucleotides over 200 is associated with the onset of X-linked mental retardation and it’s coupled with cytosine methylation that results in transcriptional silencing of the protein-coding gene FMR1 and the non-protein coding AS-FMR1 and FMR4. (C-D) Spinocerebellar Ataxia type 7. Spinocerebellar Ataxia 7 is caused by a CAG repeats expansion (37- 400) located in proximity of the translation start site of Ataxin-7 protein leading to polyglutamine (polyQ) accumulation (C). Ataxin-7 gene is featured by CAG repeats flanked by CTCF- binding sites and by an alternative TSS of the antisense RNA SCAANT1 (Spinocerebellar ataxia-7 antisense noncoding transcript 1) (D). In patients affected by Ataxia-7 the expansion of trinucleotides affects the binding of CTCF that causes reduced transcription of SCAANT1 and results in higher expression of the aberrant Ataxin-7 protein (C). (E) Spinocerebellar Atxia type 8. Spinocerebellar Ataxia type 8 arises when trinucleotide CTG in the ATXN8 gene expand over 70 repeats (70- 250). Bidirectional transcription has been detected in ATNX8 gene producing protein coding gene ATXN8 and the non-proteincoding RNA ATXN8OS. CTG repeats in ATNX8 mRNA results in accumulation of polyQ proteins while CTG repeats transcribed in antisense direction (CAG) are not translated and result in the production of toxic noncoding RNAs.
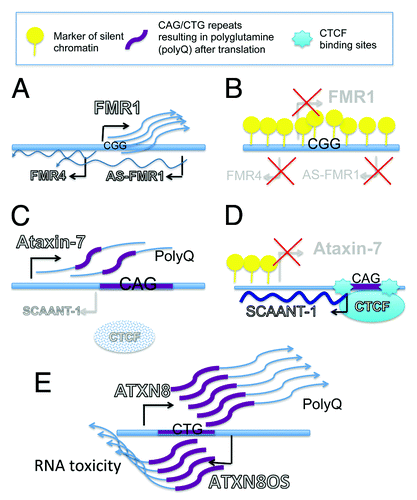
Spinocerebellar ataxias
Spinocerebellar ataxias are a diverse group of rare, slowly progressing, neurological disorders affecting the cerebellum. Spinocerebellar ataxia type 7 (SCA7) is a neurodegenerative disorder caused by CAG repeat tract (37 ≥ 400 repeats) in the ataxin-7 gene as compared with normal individuals (4–35 repeats).Citation42,Citation43 The expansion region is flanked by binding sites for the transcription factor CTCFCitation44 and by the TSS of the antisense transcript SCAANT1 (spinocerebellar ataxia 7 antisense noncoding transcript 1)Citation44 (). Evidence for CTCF binding in close proximity to the repeat sequences is emerging as a common occurrence in microsatellite expansion diseases. Importantly, CTCF binding is required for SCAANT1 transcription and it has been shown that the knockdown of SCAANT1 or the absence of CTCF increases ataxin-7 sense gene transcription. Based on this evidence, we can speculate that the overexpression of the antisense SCAANT1 could be attempted to silence the aberrant expression of Ataxin-7. CTCF and lncRNA-mediated regulation of genomic architecture could play crucial roles in transcriptional deregulation at repeat loci. Interestingly, CTCF binding sites have also been detected in the promoter region of FMR1,Citation9,Citation45 suggesting that CTCF might regulate FMR1 and FMR1 locus-related noncoding RNA transcription or regional chromatin loop formation.Citation46
Spinocerebellar ataxia type 8 (SCA8) is characterized by a nucleotide repeat expansion in a region containing two transcripts expressed in opposite directions. Nucleotide repeat involves both the ATXN8 protein coding gene and the ATXN8OS antisense noncoding gene.Citation40 Transcription of ATXN8 protein-coding RNA containing CAG repeats is translated into a polyglutamine expansion protein while the untranslated antisense partner expresses CUG repeats (). These findings suggest that SCA8 pathogenesis involves a toxic gain of function at both the RNA and protein levels. In fact, polyglutamine protein (PolyQ) inclusions have been found in SCA8 human tissuesCitation40 and CUG expansions in the antisense transcript accumulate as ribonuclear inclusions in GABAergic neurons.Citation47 Despite the known role of the antisense transcripts in causing RNA toxicity, the authors speculate on the possibility that the noncoding transcripts might have additional roles in Spinocerebellar ataxia 8 that eventually regulate sense transcript expression.Citation48
In summary, it is becoming clear that repeat expansion diseases feature: (1) genetic instability and a strong tendency for repeat expansion through generations,Citation49 (2) binding sites for CTCFCitation50 and (3) bidirectional transcription encompassing the repeats.Citation51
Huntington’s disease
Investigation of potentially important accelerators of evolution have focused primarily on protein coding genes.Citation52,Citation53 However the availability of nearly complete genome sequences for several vertebrates provides useful information to find functional elements in the roughly 99% of the genome that does not have protein coding capacity.Citation54,Citation55 Many human accelerated regions (HARs) are associated with genes involved in transcriptional regulation and neurodevelopment. The HAR1 region contains long noncoding RNAs (HAR1F and HAR1R) and is expressed early during human embryonic neocortical development.Citation27 HAR1F is specifically expressed in Cajal-Retzius neurons in the marginal zone during early development of the cortex and becomes expressed later on in other regions, e.g., in the hippocampal primordium, dentate gyrus, cerebellar cortex and olivar complex.Citation27 In the adult brain, HAR1F is expressed in the frontal cortex, hippocampus, cerebellum, thalamus and hypothalamus.Citation27 Recently, it has been shown that in the brain of patients affected by Huntington’s disease (HD), a trinucleotide expansion disease, the levels of lncRNAs HAR1F and HAR1R are decreased as compared with normal brains, suggesting a putative functional role for these lncRNAs in this disease. At the molecular level, the neurons of Huntington’s disease patients are disrupted by aberrant nuclear localization of the master transcriptional repressor REST (RE1-silencing transcription factor).Citation56 Furthermore, ChIP-seq data has revealed that the HAR1 locus is under the control of RESTCitation57 and that in the brain of HD HAR1-associated lncRNAs are part of widespread transcriptional dysfunction.
Long noncoding RNAs in imprinting-related diseases
Long noncoding RNAs have been implicated in the phenomenon of gene imprinting, where genes for only one allele (maternal or paternal) are actively transcribed. Imprinted genes are enriched in the brain and placenta and help mediate various cellular processes including cell growth and development.Citation58,Citation59 There is increasing evidence that imprinted genes are linked to neurodevelopmental processes through the regulation of cell survival, differentiation and maturation.Citation59,Citation60 Mental and neurological disorders are often influenced by deregulation of the imprinting processes. For example, genomic imprinting has been implicated in bipolar disorder, schizophrenia and autism.Citation61-Citation63 Interestingly, imprinted genes are frequently associated with antisense transcripts,Citation64 which might play a role in gene silencing and could be causal factor in neurological disorders.Citation3,Citation4
Angelman syndrome
Imprinted genes play a role in several human congenital syndromes such as Angelman syndrome (AS),Citation65 a disease characterized by severe neurologic abnormalities. The gene involved in this disorder, Ube3a, is biallelically expressed in most of the body’s tissues, however in the brain, neurons express Ube3a only from the maternal allele.Citation66 Notably, in Angelman sufferers, the maternal Ube3a gene is deleted or mutatedCitation66-Citation68 resulting in the absence of Ube3a expression in neurons. Currently, there are no effective drugs to treat this disorder, however activation of the dormant paternal allele has recently been achieved through treatment with topoisomerase inhibitors.Citation69 The activation of the silenced paternal allele is linked to a decrease in expression of the paternally expressed antisense transcript in neurons.Citation69 The mechanism behind this phenomenon remains unclear as the link between topoisomerase treatment, repression of the antisense transcript and re-expression of the gene is still obscure. These findings suggest that the re-expression of the paternal allele driven by a topoisomerase inhibitor, Topotecan, might be a potential treatment for Angelman syndrome.
Long noncoding RNAs and neuronal degeneration
Alzheimer disease
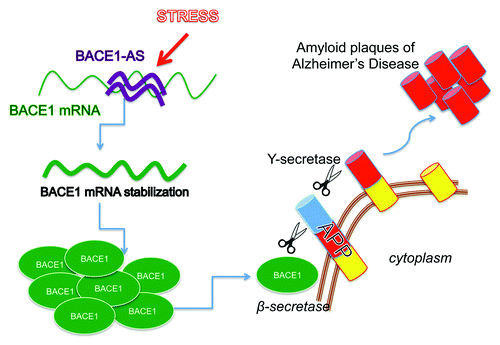
Long ncRNAs are involved in the pathogenesis of Alzheimer disease (AD),Citation14,Citation70 a neurodegenerative disorder partly characterized histologically by the deposition of amyloid plaques in the brain, due to increased levels of the β secretase enzyme (BACE1). BACE1 cleaves the amyloid precursor protein (APP) and generates amyloid peptides which can aggregate and form plaques. A conserved antisense RNA, BACE1-AS, was originally identified from data produced by large-scale transcriptomics efforts from the FANTOM consortium as one of the sense-antisense pairs conserved between human and mouse.Citation71 BACE1-AS is an approximately 2 kb RNA that is transcribed on chromosome 11, from the opposite strand of the BACE1 locus (11q 23.3). BACE1-AS shares 104 nucleotides of full complementarity to exon 6 of human BACE1 mRNA. It has been shown that BACE1-AS and BACE1 mRNA form a duplex that may act to alter the secondary or tertiary structure of BACE1 and to increase BACE1 mRNA stability.Citation14 Alzheimer disease patients display elevated levels of BACE1-AS in several brain regions as compared with controls.Citation14 Furthermore, BACE1-AS increases significantly upon cellular stress causing an upregulation in BACE1 mRNA and protein levels (). These findings are particularly interesting from a therapeutic standpoint as BACE1 remains a viable target for AD treatment.
Figure 2. BACE1-AS contributes to β -secretase increased production in the brain of Alzheimer Disease patients. Alzheimer disease associates with amyloid plaques in the brain of affected patients. The generation of these plaques derives by sequential cleavage of the amyloid precursor protein (APP) by β-secretase (BACE1) and ϒ-secretase. BACE1 is increased in affected patients due to the stabilization of the mRNA caused by the binding to its antisense noncoding RNA (BACE1-AS). Oxidative stress has been reported to cause the increase of BACE-AS in brain resulting in the increased production of β-secretase enzyme.
Long noncoding RNAs in neurologic and neuropsychiatric diseases
Certain lncRNAs have been mapped to chromosomal regions associated with neurobehavioral diseases such as schizophrenia and autism.Citation72 Many schizophrenic patients carry a translocation affecting two genes: DISC1 and DISC2 (disrupted in schizophrenia 1 and 2).Citation73 The DISC1 protein is involved in intracellular transport, cell polarity and neuronal migration and disruption of its function may in part contribute to some neurological defects.Citation28 DISC2 produces antisense non-protein coding transcripts that are transcribed from the 3′ region of protein-coding DISC1. It has been proposed that DISC2 ncRNA may be involved in the regulation of DISC1 expression.Citation73 Interestingly, it has also been suggested that the DISC1 and DISC2 genes might also play a role in the development of unipolar and bipolar depressive disorders.Citation74 The same chromosomal translocation also disrupts another ncRNA gene, PSZA11q14 (putative schizophrenia-associated gene from 11q14), which is downregulated in schizophrenia patients.Citation75 It is well recognized that neuropsychiatric conditions such as schizophrenia, depression and bipolar disorder are severe, multifactorial brain illnesses, thus it is not surprising that previously unrecognized noncoding RNAs can affect multiple pathways at different levels in these diseases.
The long ncRNA Evf-2 is transcribed from the Dlx5/6 locus and its transcription regulates that of Dlx5, Dlx6 and Gad1Citation16,Citation76 through the recruitment of transcription factors such as MeCP2. In vertebrates, Dlx genes play a critical role in neuronal developmentCitation16 and the loss of Evf2 noncoding RNA results in a decreased number of GABAergic interneurons in the early postnatal mouse hippocampus and dentate gyrus.Citation76 Schizophrenic patients show a consistent reduction of the enzyme responsible for GABA synthesis and the dysfunction of GABA-regulated circuits have been implicated in other neurological disorders such as autism and epilepsyCitation77. Thus, the discovery that Evf2 controls the development of GABAergic interneuronsCitation76 raises the possibility of targeting this pathway to attenuate specific disease-associated symptoms.
Long noncoding RNAs in brain tumors
Gliomas
Gliomas are brain tumors that originate from glial cells (astrocytes, oligodendrocytes or ependymal cells). Among these, astrocytoma grade IV (glioblastoma multiforme, GBM), is the most common malignant brain tumor in adults and is one of the deadliest cancers. Despite the development of new surgical techniques and the use of antineoplastic drugs combined with irradiation, a cure for glioblastoma does not yet exist.
Glioblastomas are a heterogeneus group of cancers that show a number of clearly defined genetic lesions. For instance, in younger patients this tumor frequently features p53 mutations. P53 is a tumor suppressor protein encoded by a gene whose disruption is associated with more than 50% percent of human cancers. The p53 protein is known as the guardian of the genome because it acts as a checkpoint in the cell cycle that either prevents or initiates programmed cell death. Recent studies identified numerous long intergenic RNAs (lincRNAs) that are induced by the p53 tumor-suppressor pathway.Citation11,Citation78 When cells are subjected to stress, lincRNAs are among the transcripts specifically induced by p53. In particular, lincRNA-p21, is directly induced by p53 and plays a critical role in the p53 transcriptional response by targeting chromatin-modifying complexes to specific genomic loci.Citation78 While p53 is known to transcriptionally activate numerous genes, the mechanisms by which p53 leads to gene repression have remained elusive. Huarte et al. propose an elegant mechanism by which p53 globally downregulates a large subset of its targets. This study emphasizes the significance of lincRNA activation in the control of transcriptional programs that maintain cellular homeostasis. Since these lincRNAs are part of p53-mediated genome surveillance, it can be argued that tumors lacking functional p53, e.g., glioblastomas, might display altered expression of lincRNAs. Another recent study identified PANDA, a lincRNA that is 5 kb upstream of CDKN1A (p21). PANDA is induced by p53 and interacts with the transcription factor NF-YA to limit the expression of pro-apoptotic genes.Citation79 Thus, the regulatory sequence upstream of CDKN1A drives the expression of multiple noncoding transcripts that coordinate the DNA damage response in healthy cells.
Inactivation, overexpression and copy number variations in the transcriptional repressor REST have been reported in several cancers. REST is a master negative regulator of neurogenesis that represses neuronal genes in non-neuronal tissues. Altered expression of REST may promote the loss of cellular identity and induce transformation through alterations in genomic stability, DNA methylation, chromatin remodeling and deregulation of onco- and tumor suppressor genes.Citation80 Abrajano et al. demonstrated that in glial cells both REST and CoREST modulate genes in key neuronal pathways.Citation80 This suggests that CoREST, like REST, might play a role in cellular transformation in the nervous system. Interestingly, the lincRNA HOTAIR serves as a scaffold for at least two distinct histone modification complexes: PRC1 (LSD1/CoREST/REST) and PRC2.Citation81 HOTAIR interaction with these distinct complexes leads to the coordinated targeting of both to chromatin. Hence, we can speculate that the deregulation of REST in brain cancers might result in epigenetic alterations depending on the delocalization of this and potentially many other lncRNAs to their target loci. A better characterization of how REST and CoREST coordinate the epigenetic programming of neural cells can help us to better understand neural development and cancer.Citation82-Citation84
The polycomb repressive complex (PRC2) catalyzes the trimethylation of histone 3 on lysine 27 (H3K27me3), to silence chromatin, altering the degree of chromatin compaction and consequently the rate of gene transcription. The mechanism by which mammalian PRC2 is recruited to chromatin is unclear. In mammals, PRC2-targeted sequences consist mostly of CpG islands, but these sequences alone do not indicate a consensus response element.Citation85 Long ncRNAs are becoming recognized as important mediators of PRC2 genomic recruitmentCitation19,Citation20,Citation23,Citation81,Citation86,Citation87 and such an interaction could be common to a large number of long ncRNAs.
Ezh2, the catalytic subunit of the PRC2 complex has been reported to interact with lncRNAs Xist,Citation86 HOTAIR,Citation88 ANRIL Citation87 and numerous lincRNAs.Citation22 Mutations in Ezh2 have been observed in lymphomaCitation89 and its overexpression is an indicator of advanced and metastatic disease in many solid tumorsCitation90 as well as gliomas, according to a recent report.Citation91 Thus, deregulation of both chromatin modifier enzymes and the lncRNAs that associate to them could easily lead to disruptions in chromatin regulation, potentially resulting in oncogenesis.
Imprinted LncRNAs and CNS tumors
It has long been suggested that chromosome 14q32 contains genes involved in the pathogenesis and progression of meningioma, a CNS tumor arising from arachnoid cells that line the brain and spinal cord. Maternally expressed gene 3 (MEG3), is an imprinted gene located on chromosome 14q32 that produces a highly expressed noncoding RNA in normal human brains that is lost in high grade meningiomas.Citation92 Interestingly, the expression of MEG3 in human meningioma cell lines strongly suppresses tumor cell growth and activates p53-mediated apoptosis,Citation92 suggesting a tumor-suppressive activity for this noncoding RNA. H19 is another example of an imprinted long noncoding RNA associated with CNS cancer. H19 is expressed during embryogenesis and the deregulation of H19 and other imprinted genes in this cluster are reported to promote cell transformation in a number of tumors, including gliomas. H19 is a target of the GLI1 transcription factor that mediates the Sonic Hedgehog signaling pathway and is amplified more than 50-fold in human gliomas.Citation93 Recently, a lncRNA antisense to H19 has been identified and its altered expression in human cancer cell lines has been reported.Citation94 The deregulation of this lncRNA in cancer further highlights the complexity of regulation within this gene cluster and may be relevant for CNS tumors.Citation30 Together, these examples demonstrate that lncRNAs are a rich source of candidate genes involved in normal health whose deregulation results in disparate diseases of the central nervous system.
Targeting of long noncoding RNAs
RNA knockdown strategies
Although the human genome is pervasively transcribed, the overwhelming majority produces non-protein coding transcripts that remain an unexplored field for drug discovery. RNA is a good candidate for drug design because it can adopt secondary structures that allow binding to specific ligands. The first RNA molecule was targeted in the late 1970sCitation95 when a short single-stranded sequence of DNA (antisense oligonucleotide, ASO), was developed to bind and block the translation of Rous sarcoma viral RNA. Since the application of the first unmodified antisense oligonucleotide molecules in 1978, numerous chemical modifications have been tested to improve the drug-like properties of ASOs. ASOs are used to knockdown genes through degradation of their respective mRNAs by RNase H (enzyme that cleaves RNA:RNA or RNA:DNA pairs) and inhibition of the translation process that does not require cleavage by RNase H. Stability, affinity and/or protein binding has been improved by the use of chemical modifications such as phosphorothioate backbones, morpholino oligomers, peptide nucleic acids (PNAs) or locked nucleic acids (LNAs). Multiple RNase H-dependent ASOs are in clinical trials, including a drug for high-grade glioma in Phase IIB trials.Citation96 The discovery of double-stranded small interfering RNAs (siRNAs) in the late 1990s added to the arsenal of RNA-targeting strategies and was successfully used in human cells a few years later.Citation97 The in vivo application of RNA interference to silence a specific transcript has shown promise but is limited by pharmacokinetic considerations and the poor ability of naked siRNAs to translocate through the cell membranes necessitating the development of novel delivery methods. Recently, we suggested a broadly applicable method to upregulate gene expression by use of AntagoNAT oligonucleotides targeted to derepress natural antisense (mostly lncRNAs) mediated inhibition (see below and ref. Citation98).
However, as with any therapeutic strategy, it is important to balance efficacy with possible adverse and off-target effects. In fact, despite encouraging results from preclinical and clinical studies and the significant progress which has been made in developing these agents as drugs, some hurdles may still exist. For example, several studies have determined that oligonucleotides can bind serum proteins affecting coagulation,Citation99 that they may cause activation of the immune systemCitation100-Citation102 and that their administration can lead to elevated transaminases, suggesting liver damage.Citation103 However, direct administration to CNS may bypass some or all of these latter potential concerns. More studies are nonetheless needed to fully assess risk profiles with centrally administered oligonucleotides.
Finally, while extremely challenging to identify, small molecules targeting RNA would arguably be the most attractive tools for regulating lncRNA activity. The interaction between small molecules and lncRNA can potentially disrupt the native conformation of the RNA, altering its activity and/or protein-binding ability. Gene therapy is a promising tool for the treatment of human diseases that cannot be cured by conventional treatments. The expression of genes epigenetically regulated by lncRNAs can be specifically modulated by targeting regulatory noncoding transcripts. This strategy has the particular advantage of inducing long lasting changes in gene expression through modulation of epigenetic events. Furthermore, targeting lncRNAs can potentially restore the expression of a silenced gene which would be out of reach for conventional drugs.
Direct delivery of RNA-targeting drugs into the CNS
The blood brain barrier (BBB) protects the brain by forming a highly selective barrier that blocks the entry of large, hydrophilic molecules. The BBB also makes delivery of drugs affecting neurologic disorders challenging. Intravenous or intraperitoneal administration of drugs usually shows very low uptake into the brain. Our group and others have pursued a strategy to bypass this obstacle through intracerebroventricularCitation104,Citation105 or intrathecal administration of oligonucleotides into the cerebrospinal fluid (CSF).Citation106-Citation111
Administration of oligonucleotides directly into the brain (interstitial, intracranial or intracerebroventricular) has indeed been fairly successful in animal models and human (). For example, an oligonucleotide with a phosphorothioate backbone targeting superoxide dismutase 1 (SOD1) was continuously infused into the right lateral ventricle of rats via a surgically implanted catheter in the skull connected to a subcutaneously embedded osmotic pump.Citation109 Treatment showed a reduction in SOD1 mRNA and protein levels in the brain and spinal cord. Mutated forms of SOD1 are responsible for familial amyotrophic lateral sclerosis (ALS), a progressive neurodegenerative disease caused by preferential loss of neurons in the upper and lower motor pathways, a disease for which current treatments are marginally beneficial. Another case where direct administration is promising is that of intracerebroventricular delivery of ASO’s to treat Alzheimer disease. Point mutations in the human APP gene in the protein region targeted by BACE1, results in familial AD. Recent work by Chauhan et al. shows that mice injected directly with antisense oligonucleotides directed at the mutated site of APP exhibit reduced βamyloid plaque deposition.Citation106
Figure 3. Administration of RNA-targeting drugs in the Central Nervous System. Several ways of administration have been developed to overcome the blood brain barrier and to deliver molecules that target the RNA in the brain. Intracranial delivery of oligonucleotides has been tested in rats and mice to target SOD1 and APP RNA to treat amyotrofic lateral sclerosis (ALS) and Alzheimer Disease (AD) respectively. Recently, intracranial delivery of antagoNATs targeting BDNF-AS showed to increase BDNF levels in mice brain suggesting a challenging application to prevent neurodegeration associated low levels of BDNF in AD. The same strategy was used to target TGF2β in a study of phase II to delivery chemoteraphy for glioblastoma (GBM). Intranasal administration of siRNA targeting the Telomerase (TERT) has been successfully used and will be potentially applied in the future to human being. Intrathecal administration, commonly used in pain relief cures, can be applied in the future for the delivery of drugs in the CSF (Blue line) to treat diseases affecting the CNS.
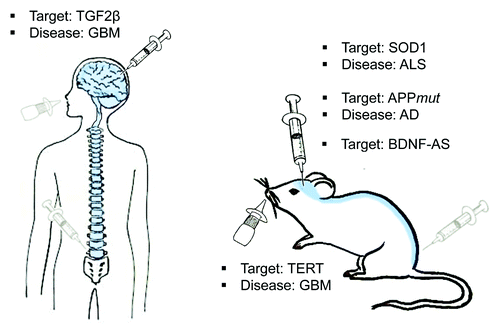
Recently, Modarresi et al., showed that the upregulation of BDNF (brain-derived neurotrophic factor) can be achieved in vivo by targeting BDNF-natural antisense transcript (BDNF-AS) with modified oligonucleotides called antagoNATs.Citation98 BDNF-AS represses the transcription of BDNF by altering the chromatin landscape at the gene locus, resulting in decreased BDNF protein levels. Oligonucleotides targeting BDNF-AS were administered to mice through intracerebroventricular delivery, using osmotic mini-pumps. After 28 d of continuous infusion of the AntagoNAT, BDNF mRNA levels were increased across forebrain regions as compared with control oligonucleotide treated mice. Importantly, the neurotrophin BDNF is a growth factor that enhances the survival, development, differentiation and function of neurons.Citation112-Citation114 Several neurodegenerative, neurodevelopmental and psychiatric disordersCitation115-Citation117 show impaired levels of neurotrophins, suggesting that the upregulation of BDNF can have beneficial effects on several disorders. It has long been known that BDNF levels are decreased in AD brainCitation118 and it also seems that the ratio of BDNF sense to BDNF antisense transcript concentration is markedly dysregulated in this disease.Citation98 In vitro and in vivo models of neurodegenerative disease show that BDNF prevents neuronal degeneration.Citation115,Citation119 Thus, its upregulation mediated by AntagoNATs might represent a potential neuroprotective agent useful to prevent neurodegeneration.
Intracranial delivery of oligonucleotides in an outpatient setting has been tested in a phase II clinical trial for human glioblastoma. Trabedersen®, an antisense oligonucleotide targeting TGF-2β, was infused intratumorally using convection-enhanced delivery (CED) via a subcutaneous port access system connected to an external pump to allow for outpatient treatment.Citation96 The treatment was safe and showed significant benefits compared with standard chemotherapy. The advantages of ventricular infusion with an implanted pump includes accurate continuous accurate deliveryCitation120 and trapping of oligonucleotides inside the brain by the BBB.Citation121
Intranasal delivery is a promising alternative to standard delivery systems as it provides a practical, painless method for delivering therapeutic agents to the brain (). Rats bearing brain tumors were treated intranasally with an oligonucleotide against Telomerase (TERT).Citation122 The intranasally delivered treatment bypassed the blood-brain barrier, reached the tumor and inhibited tumor growth, leading to a prolonged lifespan in treated rats. This delivery approach appears to selectively kill tumor cells as toxic effects were not observed in normal brain tissues. These data support further research into the intranasal delivery of tumor-specific therapeutic agents for brain cancer patients.
Finally, while intrathecal administration (IT) of drugs is well established in anesthesia and pain management, recent data suggest that oligonucleotides introduced IT can be distributed to the brain through CSF suggesting an additional route of administration to treat CNS diseases.
As discussed previously in this review, the onset of many diseases affecting the CNS occurs during development and treatment thus would ideally have to begin in the very early stages of the disease. Neverthess, Michalon et al.,Citation123 for example, have published encouraging results showing that the early and chronic inhibition of the glutamate receptor mGlu5 in young FXS mouse model can “correct” a broad range of complex behavioral and neuroanatomical defects associated to that disease. Importantly, the treatment was started in these mice at an age of 4–5 weeks, when the brain development is anatomically complete but highly plastic and when all behavioral and anatomical FXS phenotypes are evident.Citation123 These results may demonstrate the feasibility of treatments for developmental disorders, but there are a few important considerations in the design of therapies for these conditions. First, as in Michalon et al. study,Citation123 treatment must be initiated early in the course of disease, which would generally imply very young pediatric patients. In addition, as most neurodevelopmental disorders have a strong genetic component, they can rarely be completely cured and patients would therefore require long-term treatment. Because of this, therapies and delivery strategies such as those discussed in this article should be considered for potential use over the course of many years.
Biological carriers for RNA-targeting therapies
Viruses are natural vehicles for the efficient transfer of genes into host cells. This characteristic makes viruses an attractive system for the delivery of therapeutic nucleic acids. Viral vectors that have recently been used in the laboratory and in clinical practice include adenoviruses, retroviruses and lentiviruses. Non-viral vectors have also been popular tools for gene delivery, because they are relatively safe and can be modified through the incorporation of ligands to target specific cell types.Citation124 This method of delivery includes three major vectors: polymer-, lipid- and peptide-based systems. However, the levels of gene expression mediated by these vectors are low as compared with viral vectors. A new twist on conventional viral-delivery systems is the virosome. Using virosomes, vesicles can be made from reconstituted virion-like phospholipid bilayers and unlike conventional liposomes, they are able to carry various drugs including nucleic acids.Citation125 Despite their low toxicity, in vivo application of this technology is sometimes limited by the risk of immunogenicity; to overcome this obstacle virosomes have been modified with polyethylene glycol (PEG).
Carrier-based strategies are currently being developed using organisms such as bacteria that can naturally evade the host immune system to enter the target cell.Citation126 Non-pathogenic bacteria such as Lactococcus lactis or Streptococcus gordonii could serve as vehicles for the production and delivery of biologically active proteins such as cytokines, enzymes or even siRNAs. To date, recombinant Salmonella strains have been engineered to deliver siRNAs to tumors to suppress tumor growth in vivo.Citation127,Citation128 In fact, Gram-negative bacteria such as Salmonella can colonize and grow within aerobic and anaerobic areas of tumors. The potential to selectively colonize hypoxic areas of tumors that cannot be treated by chemotherapy presents an exciting opportunity for cancer therapeutics.
Another strategy involves the use of the envelope from Gram-negative bacteria that is lysed with protein E,Citation129 to produce a “bacterial ghost.” Despite the absence of the cytoplasmic contents, the surface enables encapsulation of a range of cargos and provides an intrinsic mechanism for targeting dendritic cells, macrophages and tumor cellsCitation130-Citation133
Essential components of the immune system such as macrophages can potentially be engineered to deliver therapeutic molecules due to their tendency to phagocytose nanoparticles and to target diseased tissues.Citation134,Citation135 Advances in the field of nanotechnology have led to preclinical studies in which ex-vivo-made nanoparticle-bearing macrophages are re-injected into the body as “Trojan horse” delivery carriers.Citation136 In this context, the formation of siRNA and/or oligonucleotide nanoparticles is a interesting strategy for systemic nucleic acid delivery.
Conclusions
It is becoming abundantly clear that focusing drug discovery efforts solely on conventional protein targets may not produce the therapeutics needed urgently to combat CNS disorders. The last decade has yielded much insight into the important functional role of noncoding RNAs as well as the mechanisms of action of these transcripts. The direct involvement of long noncoding RNAs in the regulation of basic biological processes leads us to hypothesize that their altered expression and/or function can have profound effects on multiple cellular pathways and that they may constitute a large new class of viable biomarker and drug targets.
Acknowledgment
The authors are grateful to all colleagues who gave helpful comments and proofread the manuscript. Our work on long noncoding RNA mechanisms in CNS disorders is in part funded by the US. National Institutes of Health (5R01NS063974 and 5R01MH084880).
References
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, et al. Divergent transcription from active promoters. Science 2008; 322:1849 - 51; http://dx.doi.org/10.1126/science.1162253; PMID: 19056940
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science 2008; 322:1851 - 4; http://dx.doi.org/10.1126/science.1164096; PMID: 19056938
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 2008; 32:232 - 46; http://dx.doi.org/10.1016/j.molcel.2008.08.022; PMID: 18951091
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 2008; 322:1717 - 20; http://dx.doi.org/10.1126/science.1163802; PMID: 18988810
- He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science 2008; 322:1855 - 7; http://dx.doi.org/10.1126/science.1163853; PMID: 19056939
- Finocchiaro G, Carro MS, Francois S, Parise P, DiNinni V, Muller H. Localizing hotspots of antisense transcription. Nucleic Acids Res 2007; 35:1488 - 500; http://dx.doi.org/10.1093/nar/gkm027; PMID: 17284453
- Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science 2008; 319:1791 - 2; http://dx.doi.org/10.1126/science.1150843; PMID: 18369138
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007; 316:1484 - 8; http://dx.doi.org/10.1126/science.1138341; PMID: 17510325
- Ladd PD, Smith LE, Rabaia NA, Moore JM, Georges SA, Hansen RS, et al. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet 2007; 16:3174 - 87; http://dx.doi.org/10.1093/hmg/ddm293; PMID: 17921506
- Khalil AM, Faghihi MA, Modarresi F, Brothers SP, Wahlestedt C. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS One 2008; 3:e1486; http://dx.doi.org/10.1371/journal.pone.0001486; PMID: 18213394
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009; 458:223 - 7; http://dx.doi.org/10.1038/nature07672; PMID: 19182780
- Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res 2011; 39:Database issue D146 - 51; http://dx.doi.org/10.1093/nar/gkq1138; PMID: 21112873
- Munroe SH, Lazar MA. Inhibition of c-erbA mRNA splicing by a naturally occurring antisense RNA. J Biol Chem 1991; 266:22083 - 6; PMID: 1657988
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med 2008; 14:723 - 30; http://dx.doi.org/10.1038/nm1784; PMID: 18587408
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009; 10:155 - 9; http://dx.doi.org/10.1038/nrg2521; PMID: 19188922
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev 2006; 20:1470 - 84; http://dx.doi.org/10.1101/gad.1416106; PMID: 16705037
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 2007; 445:666 - 70; http://dx.doi.org/10.1038/nature05519; PMID: 17237763
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 2008; 454:126 - 30; http://dx.doi.org/10.1038/nature06992; PMID: 18509338
- Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 2008; 451:202 - 6; http://dx.doi.org/10.1038/nature06468; PMID: 18185590
- Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 2010; 38:662 - 74; http://dx.doi.org/10.1016/j.molcel.2010.03.021; PMID: 20541999
- Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet 2008; 4:e1000258; http://dx.doi.org/10.1371/journal.pgen.1000258; PMID: 19008947
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 2009; 106:11667 - 72; http://dx.doi.org/10.1073/pnas.0904715106; PMID: 19571010
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007; 129:1311 - 23; http://dx.doi.org/10.1016/j.cell.2007.05.022; PMID: 17604720
- Gräff J, Kim D, Dobbin MM, Tsai LH. Epigenetic regulation of gene expression in physiological and pathological brain processes. Physiol Rev 2011; 91:603 - 49; http://dx.doi.org/10.1152/physrev.00012.2010; PMID: 21527733
- Mercer TR, Dinger ME, Mariani J, Kosik KS, Mehler MF, Mattick JS. Noncoding RNAs in Long-Term Memory Formation. Neuroscientist 2008; 14:434 - 45; http://dx.doi.org/10.1177/1073858408319187; PMID: 18997122
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J 2010; 29:3082 - 93; http://dx.doi.org/10.1038/emboj.2010.199; PMID: 20729808
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature 2006; 443:167 - 72; http://dx.doi.org/10.1038/nature05113; PMID: 16915236
- Mehler MF, Mattick JS. Non-coding RNAs in the nervous system. J Physiol 2006; 575:333 - 41; http://dx.doi.org/10.1113/jphysiol.2006.113191; PMID: 16809366
- Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, et al. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci 2010; 11:14; http://dx.doi.org/10.1186/1471-2202-11-14; PMID: 20137068
- Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res 2010; 1338:20 - 35; http://dx.doi.org/10.1016/j.brainres.2010.03.110; PMID: 20380817
- Mehler MF, Mattick JS. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol Rev 2007; 87:799 - 823; http://dx.doi.org/10.1152/physrev.00036.2006; PMID: 17615389
- Mehler MF. Epigenetics and the nervous system. Ann Neurol 2008; 64:602 - 17; http://dx.doi.org/10.1002/ana.21595; PMID: 19107999
- Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol 2010; 220:126 - 39; http://dx.doi.org/10.1002/path.2638; PMID: 19882673
- Eacker SM, Dawson TM, Dawson VL. Understanding microRNAs in neurodegeneration. Nat Rev Neurosci 2009; 10:837 - 41; http://dx.doi.org/10.1038/nrn2726; PMID: 19904280
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011; 12:861 - 74; http://dx.doi.org/10.1038/nrg3074; PMID: 22094949
- Kosik KS. The neuronal microRNA system. Nat Rev Neurosci 2006; 7:911 - 20; http://dx.doi.org/10.1038/nrn2037; PMID: 17115073
- Miller BH, Wahlestedt C. MicroRNA dysregulation in psychiatric disease. Brain Res 2010; 1338:89 - 99; http://dx.doi.org/10.1016/j.brainres.2010.03.035; PMID: 20303342
- López Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol 2010; 11:165 - 70; http://dx.doi.org/10.1038/nrm2854; PMID: 20177394
- Mirkin SM. Expandable DNA repeats and human disease. Nature 2007; 447:932 - 40; http://dx.doi.org/10.1038/nature05977; PMID: 17581576
- Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet 2006; 38:758 - 69; http://dx.doi.org/10.1038/ng1827; PMID: 16804541
- Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, Tapscott SJ. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell 2005; 20:483 - 9; http://dx.doi.org/10.1016/j.molcel.2005.09.002; PMID: 16285929
- David G, Abbas N, Stevanin G, Dürr A, Yvert G, Cancel G, et al. Cloning of the SCA7 gene reveals a highly unstable CAG repeat expansion. Nat Genet 1997; 17:65 - 70; http://dx.doi.org/10.1038/ng0997-65; PMID: 9288099
- Lindblad K, Savontaus ML, Stevanin G, Holmberg M, Digre K, Zander C, et al. An expanded CAG repeat sequence in spinocerebellar ataxia type 7. Genome Res 1996; 6:965 - 71; http://dx.doi.org/10.1101/gr.6.10.965; PMID: 8908515
- Sopher BL, Ladd PD, Pineda VV, Libby RT, Sunkin SM, Hurley JB, et al. CTCF regulates ataxin-7 expression through promotion of a convergently transcribed, antisense noncoding RNA. Neuron 2011; 70:1071 - 84; http://dx.doi.org/10.1016/j.neuron.2011.05.027; PMID: 21689595
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell 2007; 129:823 - 37; http://dx.doi.org/10.1016/j.cell.2007.05.009; PMID: 17512414
- Filippova GN. Genetics and epigenetics of the multifunctional protein CTCF. Curr Top Dev Biol 2008; 80:337 - 60; http://dx.doi.org/10.1016/S0070-2153(07)80009-3; PMID: 17950379
- Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, et al. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet 2009; 5:e1000600; http://dx.doi.org/10.1371/journal.pgen.1000600; PMID: 19680539
- Chen IC, Lin HY, Lee GC, Kao SH, Chen CM, Wu YR, et al. Spinocerebellar ataxia type 8 larger triplet expansion alters histone modification and induces RNA foci. BMC Mol Biol 2009; 10:9; http://dx.doi.org/10.1186/1471-2199-10-9; PMID: 19203395
- Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet 2005; 6:729 - 42; http://dx.doi.org/10.1038/nrg1689; PMID: 16205713
- Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet 2001; 17:520 - 7; http://dx.doi.org/10.1016/S0168-9525(01)02366-6; PMID: 11525835
- Batra R, Charizanis K, Swanson MS. Partners in crime: bidirectional transcription in unstable microsatellite disease. Hum Mol Genet 2010; 19:R1 R77 - 82; http://dx.doi.org/10.1093/hmg/ddq132; PMID: 20368264
- Clark AG, Glanowski S, Nielsen R, Thomas PD, Kejariwal A, Todd MA, et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science 2003; 302:1960 - 3; http://dx.doi.org/10.1126/science.1088821; PMID: 14671302
- Nielsen R. Molecular signatures of natural selection. Annu Rev Genet 2005; 39:197 - 218; http://dx.doi.org/10.1146/annurev.genet.39.073003.112420; PMID: 16285858
- Lander ES. Initial impact of the sequencing of the human genome. Nature 2011; 470:187 - 97; http://dx.doi.org/10.1038/nature09792; PMID: 21307931
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 2005; 15:1034 - 50; http://dx.doi.org/10.1101/gr.3715005; PMID: 16024819
- Zuccato C, Cattaneo E. Role of brain-derived neurotrophic factor in Huntington’s disease. Prog Neurobiol 2007; 81:294 - 330; http://dx.doi.org/10.1016/j.pneurobio.2007.01.003; PMID: 17379385
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science 2007; 316:1497 - 502; http://dx.doi.org/10.1126/science.1141319; PMID: 17540862
- Fowden AL, Sibley C, Reik W, Constancia M. Imprinted genes, placental development and fetal growth. Horm Res 2006; 65:Suppl 3 50 - 8; http://dx.doi.org/10.1159/000091506; PMID: 16612114
- Davies W, Isles AR, Wilkinson LS. Imprinted gene expression in the brain. Neurosci Biobehav Rev 2005; 29:421 - 30; http://dx.doi.org/10.1016/j.neubiorev.2004.11.007; PMID: 15820547
- Wilkinson LS, Davies W, Isles AR. Genomic imprinting effects on brain development and function. Nat Rev Neurosci 2007; 8:832 - 43; http://dx.doi.org/10.1038/nrn2235; PMID: 17925812
- Schulze TG, Buervenich S, Badner JA, Steele CJ, Detera-Wadleigh SD, Dick D, et al. Loci on chromosomes 6q and 6p interact to increase susceptibility to bipolar affective disorder in the national institute of mental health genetics initiative pedigrees. Biol Psychiatry 2004; 56:18 - 23; http://dx.doi.org/10.1016/j.biopsych.2004.04.004; PMID: 15219468
- Schanen NC. Epigenetics of autism spectrum disorders. Hum Mol Genet 2006; 15:Spec No 2 R138 - 50; http://dx.doi.org/10.1093/hmg/ddl213; PMID: 16987877
- Francks C, DeLisi LE, Shaw SH, Fisher SE, Richardson AJ, Stein JF, et al. Parent-of-origin effects on handedness and schizophrenia susceptibility on chromosome 2p12-q11. Hum Mol Genet 2003; 12:3225 - 30; http://dx.doi.org/10.1093/hmg/ddg362; PMID: 14583442
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al, RIKEN Genome Exploration Research Group, Genome Science Group (Genome Network Project Core Group), FANTOM Consortium. Antisense transcription in the mammalian transcriptome. Science 2005; 309:1564 - 6; http://dx.doi.org/10.1126/science.1112009; PMID: 16141073
- Chamberlain SJ, Lalande M. Angelman syndrome, a genomic imprinting disorder of the brain. J Neurosci 2010; 30:9958 - 63; http://dx.doi.org/10.1523/JNEUROSCI.1728-10.2010; PMID: 20668179
- Yamasaki K, Joh K, Ohta T, Masuzaki H, Ishimaru T, Mukai T, et al. Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Hum Mol Genet 2003; 12:837 - 47; http://dx.doi.org/10.1093/hmg/ddg106; PMID: 12668607
- Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet 1997; 15:70 - 3; http://dx.doi.org/10.1038/ng0197-70; PMID: 8988171
- Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, et al. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet 1997; 15:74 - 7; http://dx.doi.org/10.1038/ng0197-74; PMID: 8988172
- Huang HS, Allen JA, Mabb AM, King IF, Miriyala J, Taylor-Blake B, et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature 2011; 481:185 - 9; http://dx.doi.org/10.1038/nature10726; PMID: 22190039
- Modarresi F, Faghihi MA, Patel NS, Sahagan BG, Wahlestedt C, Lopez-Toledano MA. Knockdown of BACE1-AS Nonprotein-Coding Transcript Modulates Beta-Amyloid-Related Hippocampal Neurogenesis. Int J Alzheimers Dis 2011; 2011:929042; PMID: 21785702
- Engström PG, Suzuki H, Ninomiya N, Akalin A, Sessa L, Lavorgna G, et al. Complex Loci in human and mouse genomes. PLoS Genet 2006; 2:e47; http://dx.doi.org/10.1371/journal.pgen.0020047; PMID: 16683030
- Vincent JB, Petek E, Thevarkunnel S, Kolozsvari D, Cheung J, Patel M, et al. The RAY1/ST7 tumor-suppressor locus on chromosome 7q31 represents a complex multi-transcript system. Genomics 2002; 80:283 - 94; http://dx.doi.org/10.1006/geno.2002.6835; PMID: 12213198
- Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet 2000; 9:1415 - 23; http://dx.doi.org/10.1093/hmg/9.9.1415; PMID: 10814723
- Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders--cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet 2001; 69:428 - 33; http://dx.doi.org/10.1086/321969; PMID: 11443544
- Sokolov BP, Polesskaya OO, Uhl GR. Mouse brain gene expression changes after acute and chronic amphetamine. J Neurochem 2003; 84:244 - 52; http://dx.doi.org/10.1046/j.1471-4159.2003.01523.x; PMID: 12558987
- Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci 2009; 12:1020 - 7; http://dx.doi.org/10.1038/nn.2371; PMID: 19620975
- Di Cristo G, Pizzorusso T, Cancedda L, Sernagor E. GABAergic circuit development and its implication for CNS disorders. Neural Plast 2011; 2011:623705; http://dx.doi.org/10.1155/2011/623705; PMID: 22013542
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010; 142:409 - 19; http://dx.doi.org/10.1016/j.cell.2010.06.040; PMID: 20673990
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 2011; 43:621 - 9; http://dx.doi.org/10.1038/ng.848; PMID: 21642992
- Abrajano JJ, Qureshi IA, Gokhan S, Zheng D, Bergman A, Mehler MF. Differential deployment of REST and CoREST promotes glial subtype specification and oligodendrocyte lineage maturation. PLoS One 2009; 4:e7665; http://dx.doi.org/10.1371/journal.pone.0007665; PMID: 19888342
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010; 329:689 - 93; http://dx.doi.org/10.1126/science.1192002; PMID: 20616235
- Majumder S. REST in good times and bad: roles in tumor suppressor and oncogenic activities. Cell Cycle 2006; 5:1929 - 35; http://dx.doi.org/10.4161/cc.5.17.2982; PMID: 16929174
- Lawinger P, Venugopal R, Guo ZS, Immaneni A, Sengupta D, Lu W, et al. The neuronal repressor REST/NRSF is an essential regulator in medulloblastoma cells. Nat Med 2000; 6:826 - 31; http://dx.doi.org/10.1038/77565; PMID: 10888935
- Coulson JM. Transcriptional regulation: cancer, neurons and the REST. Curr Biol 2005; 15:R665 - 8; http://dx.doi.org/10.1016/j.cub.2005.08.032; PMID: 16139198
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 2008; 4:e1000242; http://dx.doi.org/10.1371/journal.pgen.1000242; PMID: 18974828
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008; 322:750 - 6; http://dx.doi.org/10.1126/science.1163045; PMID: 18974356
- Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 2011; 30:1956 - 62; http://dx.doi.org/10.1038/onc.2010.568; PMID: 21151178
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464:1071 - 6; http://dx.doi.org/10.1038/nature08975; PMID: 20393566
- Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet 2010; 42:722 - 6; http://dx.doi.org/10.1038/ng.621; PMID: 20601953
- Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res 2011; 17:2613 - 8; http://dx.doi.org/10.1158/1078-0432.CCR-10-2156; PMID: 21367748
- Orzan F, Pellegatta S, Poliani PL, Pisati F, Caldera V, Menghi F, et al. Enhancer of Zeste 2 (EZH2) is up-regulated in malignant gliomas and in glioma stem-like cells. Neuropathol Appl Neurobiol 2011; 37:381 - 94; http://dx.doi.org/10.1111/j.1365-2990.2010.01132.x; PMID: 20946108
- Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR, et al. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab 2003; 88:5119 - 26; http://dx.doi.org/10.1210/jc.2003-030222; PMID: 14602737
- Kinzler KW, Bigner SH, Bigner DD, Trent JM, Law ML, O’Brien SJ, et al. Identification of an amplified, highly expressed gene in a human glioma. Science 1987; 236:70 - 3; http://dx.doi.org/10.1126/science.3563490; PMID: 3563490
- Berteaux N, Aptel N, Cathala G, Genton C, Coll J, Daccache A, et al. A novel H19 antisense RNA overexpressed in breast cancer contributes to paternal IGF2 expression. Mol Cell Biol 2008; 28:6731 - 45; http://dx.doi.org/10.1128/MCB.02103-07; PMID: 18794369
- Zamecnik PC, Stephenson ML. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc Natl Acad Sci U S A 1978; 75:280 - 4; http://dx.doi.org/10.1073/pnas.75.1.280; PMID: 75545
- Bogdahn U, Hau P, Stockhammer G, Venkataramana NK, Mahapatra AK, Suri A, et al, Trabedersen Glioma Study Group. Targeted therapy for high-grade glioma with the TGF-β2 inhibitor trabedersen: results of a randomized and controlled phase IIb study. Neuro Oncol 2011; 13:132 - 42; http://dx.doi.org/10.1093/neuonc/noq142; PMID: 20980335
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001; 411:494 - 8; http://dx.doi.org/10.1038/35078107; PMID: 11373684
- Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol 2012; In press http://dx.doi.org/10.1038/nbt.2158; PMID: 22446693
- Henry SP, Novotny W, Leeds J, Auletta C, Kornbrust DJ. Inhibition of coagulation by a phosphorothioate oligonucleotide. Antisense Nucleic Acid Drug Dev 1997; 7:503 - 10; http://dx.doi.org/10.1089/oli.1.1997.7.503; PMID: 9361909
- Krieg AM, Yi AK, Matson S, Waldschmidt TJ, Bishop GA, Teasdale R, et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 1995; 374:546 - 9; http://dx.doi.org/10.1038/374546a0; PMID: 7700380
- Liang H, Nishioka Y, Reich CF, Pisetsky DS, Lipsky PE. Activation of human B cells by phosphorothioate oligodeoxynucleotides. J Clin Invest 1996; 98:1119 - 29; http://dx.doi.org/10.1172/JCI118894; PMID: 8787674
- Prasad V, Hashim S, Mukhopadhyay A, Basu SK, Roy RP. Oligonucleotides tethered to a short polyguanylic acid stretch are targeted to macrophages: enhanced antiviral activity of a vesicular stomatitis virus-specific antisense oligonucleotide. Antimicrob Agents Chemother 1999; 43:2689 - 96; PMID: 10543748
- Agrawal S, Zhao Q, Jiang Z, Oliver C, Giles H, Heath J, et al. Toxicologic effects of an oligodeoxynucleotide phosphorothioate and its analogs following intravenous administration in rats. Antisense Nucleic Acid Drug Dev 1997; 7:575 - 84; http://dx.doi.org/10.1089/oli.1.1997.7.575; PMID: 9450915
- Wahlestedt C, Golanov E, Yamamoto S, Yee F, Ericson H, Yoo H, et al. Antisense oligodeoxynucleotides to NMDA-R1 receptor channel protect cortical neurons from excitotoxicity and reduce focal ischaemic infarctions. Nature 1993; 363:260 - 3; http://dx.doi.org/10.1038/363260a0; PMID: 8487863
- Wahlestedt C, Pich EM, Koob GF, Yee F, Heilig M. Modulation of anxiety and neuropeptide Y-Y1 receptors by antisense oligodeoxynucleotides. Science 1993; 259:528 - 31; http://dx.doi.org/10.1126/science.8380941; PMID: 8380941
- Chauhan NB, Siegel GJ. Antisense inhibition at the beta-secretase-site of beta-amyloid precursor protein reduces cerebral amyloid and acetyl cholinesterase activity in Tg2576. Neuroscience 2007; 146:143 - 51; http://dx.doi.org/10.1016/j.neuroscience.2007.01.008; PMID: 17303345
- Luo MC, Zhang DQ, Ma SW, Huang YY, Shuster SJ, Porreca F, et al. An efficient intrathecal delivery of small interfering RNA to the spinal cord and peripheral neurons. Mol Pain 2005; 1:29; http://dx.doi.org/10.1186/1744-8069-1-29; PMID: 16191203
- Passini MA, Bu J, Richards AM, Kinnecom C, Sardi SP, Stanek LM, et al. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci Transl Med 2011; 3:72ra18; http://dx.doi.org/10.1126/scitranslmed.3001777; PMID: 21368223
- Smith RA, Miller TM, Yamanaka K, Monia BP, Condon TP, Hung G, et al. Antisense oligonucleotide therapy for neurodegenerative disease. J Clin Invest 2006; 116:2290 - 6; http://dx.doi.org/10.1172/JCI25424; PMID: 16878173
- Standifer KM, Chien CC, Wahlestedt C, Brown GP, Pasternak GW. Selective loss of delta opioid analgesia and binding by antisense oligodeoxynucleotides to a delta opioid receptor. Neuron 1994; 12:805 - 10; http://dx.doi.org/10.1016/0896-6273(94)90333-6; PMID: 8161452
- Thakker DR, Natt F, Hüsken D, Maier R, Müller M, van der Putten H, et al. Neurochemical and behavioral consequences of widespread gene knockdown in the adult mouse brain by using nonviral RNA interference. Proc Natl Acad Sci U S A 2004; 101:17270 - 5; http://dx.doi.org/10.1073/pnas.0406214101; PMID: 15569935
- Hasbi A, Fan T, Alijaniaram M, Nguyen T, Perreault ML, O’Dowd BF, et al. Calcium signaling cascade links dopamine D1-D2 receptor heteromer to striatal BDNF production and neuronal growth. Proc Natl Acad Sci U S A 2009; 106:21377 - 82; http://dx.doi.org/10.1073/pnas.0903676106; PMID: 19948956
- Choi DC, Maguschak KA, Ye K, Jang SW, Myers KM, Ressler KJ. Prelimbic cortical BDNF is required for memory of learned fear but not extinction or innate fear. Proc Natl Acad Sci U S A 2010; 107:2675 - 80; http://dx.doi.org/10.1073/pnas.0909359107; PMID: 20133801
- Antal A, Chaieb L, Moliadze V, Monte-Silva K, Poreisz C, Thirugnanasambandam N, et al. Brain-derived neurotrophic factor (BDNF) gene polymorphisms shape cortical plasticity in humans. Brain Stimul 2010; 3:230 - 7; http://dx.doi.org/10.1016/j.brs.2009.12.003; PMID: 20965453
- Massa SM, Yang T, Xie Y, Shi J, Bilgen M, Joyce JN, et al. Small molecule BDNF mimetics activate TrkB signaling and prevent neuronal degeneration in rodents. J Clin Invest 2010; 120:1774 - 85; http://dx.doi.org/10.1172/JCI41356; PMID: 20407211
- Laske C, Stellos K, Hoffmann N, Stransky E, Straten G, Eschweiler GW, et al. Higher BDNF serum levels predict slower cognitive decline in Alzheimer’s disease patients. Int J Neuropsychopharmacol 2011; 14:399 - 404; http://dx.doi.org/10.1017/S1461145710001008; PMID: 20860877
- Dell’Osso L, Del Debbio A, Veltri A, Bianchi C, Roncaglia I, Carlini M, et al. Associations between brain-derived neurotrophic factor plasma levels and severity of the illness, recurrence and symptoms in depressed patients. Neuropsychobiology 2010; 62:207 - 12; http://dx.doi.org/10.1159/000319946; PMID: 20714169
- Phillips HS, Hains JM, Armanini M, Laramee GR, Johnson SA, Winslow JW. BDNF mRNA is decreased in the hippocampus of individuals with Alzheimer’s disease. Neuron 1991; 7:695 - 702; http://dx.doi.org/10.1016/0896-6273(91)90273-3; PMID: 1742020
- Nagahara AH, Merrill DA, Coppola G, Tsukada S, Schroeder BE, Shaked GM, et al. Neuroprotective effects of brain-derived neurotrophic factor in rodent and primate models of Alzheimer’s disease. Nat Med 2009; 15:331 - 7; http://dx.doi.org/10.1038/nm.1912; PMID: 19198615
- Dash AK, Cudworth GC 2nd. Therapeutic applications of implantable drug delivery systems. J Pharmacol Toxicol Methods 1998; 40:1 - 12; http://dx.doi.org/10.1016/S1056-8719(98)00027-6; PMID: 9920528
- Hua Y, Sahashi K, Hung G, Rigo F, Passini MA, Bennett CF, et al. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev 2010; 24:1634 - 44; http://dx.doi.org/10.1101/gad.1941310; PMID: 20624852
- Hashizume R, Ozawa T, Gryaznov SM, Bollen AW, Lamborn KR, Frey WH 2nd, et al. New therapeutic approach for brain tumors: Intranasal delivery of telomerase inhibitor GRN163. Neuro Oncol 2008; 10:112 - 20; http://dx.doi.org/10.1215/15228517-2007-052; PMID: 18287341
- Michalon A, Sidorov M, Ballard TM, Ozmen L, Spooren W, Wettstein JG, et al. Chronic Pharmacological mGlu5 Inhibition Corrects Fragile X in Adult Mice. Neuron 2012; 74:49 - 56; PMID: 22500629
- Waehler R, Russell SJ, Curiel DT. Engineering targeted viral vectors for gene therapy. Nat Rev Genet 2007; 8:573 - 87; http://dx.doi.org/10.1038/nrg2141; PMID: 17607305
- Daemen T, de Mare A, Bungener L, de Jonge J, Huckriede A, Wilschut J. Virosomes for antigen and DNA delivery. Adv Drug Deliv Rev 2005; 57:451 - 63; http://dx.doi.org/10.1016/j.addr.2004.09.005; PMID: 15560951
- Ploegh HL. Viral strategies of immune evasion. Science 1998; 280:248 - 53; http://dx.doi.org/10.1126/science.280.5361.248; PMID: 9535648
- Jiang Z, Zhao P, Zhou Z, Liu J, Qin L, Wang H. Using attenuated Salmonella typhi as tumor targeting vector for MDR1 siRNA delivery. Cancer Biol Ther 2007; 6:555 - 60; http://dx.doi.org/10.4161/cbt.6.4.3850; PMID: 17374987
- Zhang L, Ambulos N, Mixson AJ. DNA delivery to cells in culture using peptides. Methods Mol Biol 2004; 245:33 - 52; PMID: 14707368
- Witte A, Wanner G, Sulzner M, Lubitz W. Dynamics of PhiX174 protein E-mediated lysis of Escherichia coli. Arch Microbiol 1992; 157:381 - 8; http://dx.doi.org/10.1007/BF00248685; PMID: 1534215
- Kudela P, Paukner S, Mayr UB, Cholujova D, Schwarczova Z, Sedlak J, et al. Bacterial ghosts as novel efficient targeting vehicles for DNA delivery to the human monocyte-derived dendritic cells. J Immunother 2005; 28:136 - 43; http://dx.doi.org/10.1097/01.cji.0000154246.89630.6f; PMID: 15725957
- Paukner S, Kohl G, Lubitz W. Bacterial ghosts as novel advanced drug delivery systems: antiproliferative activity of loaded doxorubicin in human Caco-2 cells. J Control Release 2004; 94:63 - 74; http://dx.doi.org/10.1016/j.jconrel.2003.09.010; PMID: 14684272
- Paukner S, Kudela P, Kohl G, Schlapp T, Friedrichs S, Lubitz W. DNA-loaded bacterial ghosts efficiently mediate reporter gene transfer and expression in macrophages. Mol Ther 2005; 11:215 - 23; http://dx.doi.org/10.1016/j.ymthe.2004.09.024; PMID: 15668133
- Lubitz P, Mayr UB, Lubitz W. Applications of bacterial ghosts in biomedicine. Adv Exp Med Biol 2009; 655:159 - 70; http://dx.doi.org/10.1007/978-1-4419-1132-2_12; PMID: 20047041
- Burke B, Tang N, Corke KP, Tazzyman D, Ameri K, Wells M, et al. Expression of HIF-1alpha by human macrophages: implications for the use of macrophages in hypoxia-regulated cancer gene therapy. J Pathol 2002; 196:204 - 12; http://dx.doi.org/10.1002/path.1029; PMID: 11793372
- Burke B, Sumner S, Maitland N, Lewis CE. Macrophages in gene therapy: cellular delivery vehicles and in vivo targets. J Leukoc Biol 2002; 72:417 - 28; PMID: 12223508
- Dou H, Grotepas CB, McMillan JM, Destache CJ, Chaubal M, Werling J, et al. Macrophage delivery of nanoformulated antiretroviral drug to the brain in a murine model of neuroAIDS. J Immunol 2009; 183:661 - 9; http://dx.doi.org/10.4049/jimmunol.0900274; PMID: 19535632