Abstract
Two thirds of the human genome is composed of repetitive sequences. Despite their prevalence, DNA repeats are largely ignored. The vast majority of our genome is transcribed to produce non protein-coding RNAs. Among these, long non protein-coding RNAs represent the most prevalent and functionally diverse class. The relevance of the non protein-coding genome to human disease has mainly been studied regarding the altered microRNA expression and function in human cancer. On the contrary, the elucidation of the involvement of long non-coding RNAs in disease is only in its infancy. We have recently found that a chromatin associated, long non protein-coding RNA regulates a Polycomb/Trithorax epigenetic switch at the basis of the repeat associated facioscapulohumeral muscular dystrophy, a common muscle disorder. Based on this, we propose that long non-coding RNAs produced by repetitive sequences contribute in shaping the epigenetic landscape in normal human physiology and in disease.
Long Non Protein-Coding RNAs
Nowadays, we are witnessing a progressive deconstruction of the protein-centric view that has governed the molecular biology field for decades.Citation1 Although functional non protein-coding RNAs (ncRNAs) such as tRNAs and snRNAs were discovered long time ago, the recognition of RNA as a functional molecule per se, acting at key levels in the complex, multilayered regulation of eukaryotic gene expression, occurred only in recent years.Citation2 This was triggered by the milestone discovery that the vast majority of mammalian genomes is transcribed to generate a wide array of ncRNAs.Citation3
The main classification of ncRNAs is based on their length. Typically, molecules shorter than 200 bp are termed short or small ncRNAs, while transcript greater than 200 bp are defined long ncRNAs (lncRNAs).Citation4
Short ncRNAs, which include microRNAs, piwi-interacting RNAs and short interfering RNAs, have been the most extensively characterized in recent years.Citation5 They are involved in several biological processes, such as post-transcriptional gene regulation, heterochromatin formation and mobile repetitive elements control.Citation5
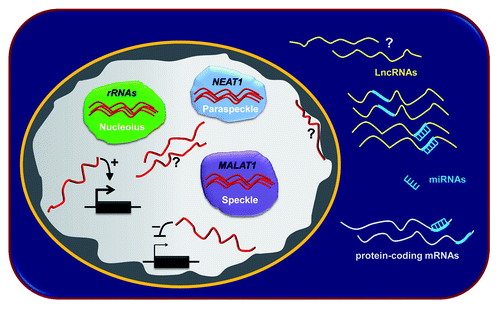
LncRNAs represent the most numerous and functionally diverse class of RNA produced by mammalian cells.Citation6 They are predominantly nuclearCitation6 and have been involved in regulation of gene expression at transcriptional and post-transcriptional levels and in the formation of functional sub-compartments in the nucleusCitation2,Citation7 (). Despite the growing interest on lncRNAs, they still remain poorly explored in terms of biological relevance, cellular function and mechanism of action.
Figure 1. Schematic representation of the diverse cellular functions of lncRNAs. LncRNAs are localized both in the cytoplasm (in yellow) and in the nucleus (in red). The vast majority of lncRNAs has yet to be functionally characterized (?). Inside the nucleus, lncRNAs can act in the negative (-) or positive (+) regulation of gene expression at transcriptional level. Furthermore, specific nuclear lncRNAs can promote a functional compartmentalization of the nucleus. They are involved in the constitution of specific sub-nuclear structures such as nucleoli (rRNAs), speckles (MALAT1) and paraspeckles (NEAT1). It has also been described that cytoplasmic lncRNAs (in yellow) can compete with mRNAs (in grey) for the binding to miRNAs (in light blue), interfering with the available cellular pool of miRNAs, ultimately influencing gene expression at the post-transcriptional level.
Most of the ncRNA analyses are performed on the non-repetitive, non-exonic, polyA+ portion of the mammalian transcriptomes. It has to be noted that over two-thirds of the mammalian genome is composed of repetitive elements.Citation8 Moreover, there are many examples of ncRNAs that are antisense or partially overlapping to protein-coding genes.Citation3 Finally, there are indications that the majority of ncRNAs are non-polyadenylated.Citation6 Hence, the range, depth and complexity of the mammalian transcriptome are far from being fully characterized.Citation9
Chromatin remodeling factors are crucial players in defining the epigenetic state of a cell and their genomic recruitment needs fine-tuned temporal and spatial regulation. Generally, chromatin remodeling components lack sequence-specific DNA-binding domains and need to be guided to their targets by auxiliary factors. Interestingly, lncRNAs tend to be enriched in the nucleus and are increasingly involved in the epigenetic regulation of gene expression.Citation6,Citation10 In particular, several lncRNAs bind to multiple chromatin regulatory proteins to mediate their recruitment to specific genomic targets.Citation11-Citation19
LncRNAs display features that are particularly suitable for molecules mediating the targeting of protein complexes to precise genomic sites. Indeed, compared to transcription factors, which typically bind to simple and degenerate DNA motifs, lncRNAs can specify “unique addresses” in the genome. Additionally, lncRNAs can form secondary structures that can function as “bait” for protein interaction, suggesting that transcription of lncRNAs might contribute to the formation of specific epigenetic landscapes.
A paradigmatic example of the role of lncRNAs in epigenetic gene regulation is represented by X-chromosome inactivation. Upon its discovery more than fifty years ago,Citation20 this has become one of the most exciting fields in molecular biology. X-chromosome inactivation is a multi-step cascade of events where several lncRNAs, produced by the so-called X-chromosome Inactivation Center (XIC), collaborate with Polycomb Group (PcG) of proteins leading to chromatin compaction and epigenetic repression.Citation21
Together with the Trithorax group (TrxG) of proteins, PcG constitutes an evolutionary conserved antagonistic system that regulates gene expression at the epigenetic level.Citation22,Citation23 Typically, PcG is associated with gene repression while TrxG with gene activation. Initially discovered as key regulators of homeotic (Hox) genes during development in D. melanogaster, it is now clear that PcG and TrxG factors play crucial roles in many biological phenomena such as cell proliferation,Citation24 stem cell identity,Citation25 cancer,Citation26 genomic imprintingCitation27 and X-chromosome inactivation.Citation28
In mammalian X-inactivation, the 1.7 kb lncRNA RepA, by directly interacting with one of its components, recruits the multiprotein complex Polycomb Repressive Complex 2 (PRC2) to the future inactive X chromosome (Xi) enabling full induction of the lncRNA Xist.Citation13,Citation29 With the help of YY1,Citation30 Polycomb Repressive Complex 1 (PRC1)Citation31 and PRC2,Citation13 Xist “paints” the X-chromosome and mediates the epigenetic repression of the whole chromosome territory. On the active X-chromosome (Xa), instead, the antisense 40 kb Tsix lncRNACitation32 functions by maintaining low Xist expression levels and by interfering with PcG binding to RepA, thus preventing the repressive cascade.Citation13,Citation33
Pioneering studies conducted in flies revealed that transcription of ncRNAs from PcG/TrxG binding sites can counteract PcG silencing.Citation11,Citation34-Citation36 The discovery in mammals of similar transcripts originating from PcG/TrxG targets such as the Hox clusterCitation15,Citation37 ,suggests that ncRNA transcription could be a general feature of the regulation of PcG/TrxG function.
So far, the exact molecular mechanism(s) through which lncRNAs recruit epigenetic regulators remains largely unclear. Moreover, the vast majority of the lncRNAs characterized up to now function in trans to epigenetically repress gene expression.
We have recently contributed to this field through the identification of the first activating lncRNA involved in a human genetic disease: facioscapulohumeral muscular dystrophy (FSHD) (see below).
Summary of Our Paper
FSHD (MIM #158900) is the third most common myopathyCitation38 and is characterized by progressive wasting of facial, upper arm and shoulder girdle muscles. FSHD is an autosomal-dominant hereditary disorder with peculiar features.Citation39 The disease is not caused by classical mutations in a protein-coding gene. Rather, it is associated with reduction in the copy number of the 3.3 kb macrosatellite D4Z4 repeat mapping to the sub-telomeric region of human chromosome 4 long arm (4q35). D4Z4 is extremely polymorphic in the general population, ranging from 11 to 150 copies.Citation40,Citation41 On the contrary, FSHD patients carry a contracted D4Z4 array, containing only 1 to 10 units,Citation42,Citation43 suggesting that a gain-of-function element, linked to a threshold D4Z4 copy number effect, is involved in the disease.
D4Z4 belongs to a family of human tandem repeats termed macrosatellites that are non-centromerically located.Citation44 Together with other members of the family, such as DXZ4 on chromosome XCitation45 and RS447 on 4p,Citation46 D4Z4 is extremely GC-rich. Notably, the area occupied by D4Z4 repeats in healthy subjects represents one of the largest GC-rich regions of the human genome.
Several FSHD clinical features, such as the variability in severity and rate of progression, the gender bias in penetrance, the asymmetric muscle wasting and the discordance of the disease in monozygotic twins, strongly suggest the involvement of epigenetic factors.Citation47 Accordingly, DNA methylation,Citation48 histone modificationsCitation49,Citation50 and higher order chromatin structureCitation49,Citation51,Citation52 are altered in FSHD patients. While in healthy subjects the 4q35 locus is organized as repressed chromatin, FSHD has been associated with an epigenetic switch that leads to the inappropriate de-repression of several 4q35 genes, among which there are the leading FSHD candidates.Citation53,Citation54 While this is known since over a decade, the molecular mechanism through which D4Z4 repeats regulate chromatin structure and gene expression at 4q35 has remained elusive.
Our recent work indicates that the D4Z4 repeat is a novel Polycomb target.Citation18 Indeed, D4Z4 repeats are able to initiate de novo PcG recruitment and a reduction of PcG levels causes de-repression of the 4q35 locus. Importantly, FSHD patients display a reduced PcG binding and a reduced spreading of the PcG histone mark H3K27me3 at the disease locus compared to controls.Citation18
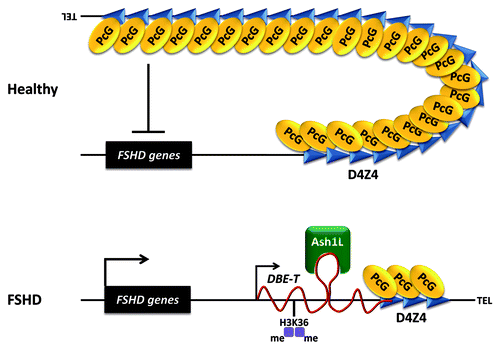
Based on our data, we propose that in healthy subjects, the presence of many D4Z4 units results in extensive PcG binding, DNA methylation, histone de-acetylation and chromatin compaction leading to a repressive chromatin organization. In FSHD patients, reduction of D4Z4 copy number results in a critical reduction of PcG binding and, as a consequence, reduction of PcG silencing on the contracted 4q35 allele. This creates the epigenetic environment permissive for the transcription of an activatory, long ncRNA, that we named DBE-T, originating proximally to and covering part of the repeat array (). DBE-T is produced solely in FSHD patients and is required for opening up the 4q35 chromatin and for de-repression of 4q35 genes. Mechanistically, we discovered that DBE-T is a chromatin-associated RNA that functions in cis, as demonstrated by the lack of 4q35 genes transcriptional de-repression in cells expressing DBE-T in trans from a transgene. Our data show that DBE-T remains associated to the FSHD locus chromatin and promotes the recruitment of the TrxG protein Ash1L through direct binding. Ash1L recruitment is associated with the accumulation of H3K36me2 at the FSHD locus. As a consequence, this leads to de-repression of FSHD candidate genesCitation18 (). In this respect, DBE-T appears a very interesting target to develop therapeutic approaches aimed at normalizing 4q35 gene expression in FSHD patients.
Figure 2. Model of DBE-T mediated de-repression of 4q35 genes in FSHD. In healthy subjects, the D4Z4 repeat array carrying many units displays extensive binding of PcG proteins leading to repression of the 4q35 locus. In FSHD patients, reduction in D4Z4 copy number critically diminishes PcG binding and silencing, allowing for transcription of the lncRNA DBE-T to occur. DBE-T functions in cis to promote opening of chromatin structure and de-repression of 4q35 genes through direct binding and recruitment of the TrxG protein Ash1L, which drives H3K36me2 at the FSHD locus.
Open Questions
Classical FSHD patients (FSHD1) carry the deletion of just D4Z4 repeats on a single 4q35 allele. FSHD1 patients carrying also deletions proximal to the D4Z4 arrays have been described.Citation55 Notably, some of these proximal deletions cover the region were we mapped the DBE-T transcriptional start site (TSS). Future work is required to determine if DBE-T (possibly a truncated version or a version starting from an alternative start site) is still produced in these patients.
A far less common form of the disease, FSHD2 (MIM #158901), is not linked to D4Z4 deletions at 4q35 but, through an unknown mechanism, is also characterized by loss of heterochromatin at the FSHD locus.Citation56 Interestingly, FSHD2 patients display loss of heterochromatin not only from 4q35 but also from 10q26, where a macrosatellite repeat array almost identical to D4Z4 exists, suggesting a functional communication between these chromosomes, reminiscent of transvection in Drosophila.Citation57 Consistent with this, somatic pairing of 4q35 and 10q26 D4Z4 has been reported.Citation58 Whether DBE-T is produced also in FSHD2 patients and has a role in the general loss of heterochromatin at 4q35 and 10q26 remains to be investigated.
The copy number of D4Z4 repeats is inversely correlated to the age of onset and clinical severity of FSHD.Citation59-Citation64 In general, alleles with 1-3 D4Z4 repeats are associated with a severe form of disease that presents in childhood, while patients with 8-10 repeats develop a milder disease with reduced penetrance. Interestingly, in FSHD the disease affects the various muscle types differently. Recently, a possible relationship between clinical and epigenetic parameters was reported.Citation56 It would be important to determine if DBE-T plays a role in mediating the epigenetic component of the clinical severity in FSHD.
A recent study conducted in yeast suggests that lncRNAs acting in cis could be responsible for the generation of a wide spectrum of fine-tuned, modulated effects on their targets, allowing for the production of variegated gene expression and, as a consequence, variegated phenotypes among genetically identical cells.Citation65 In this perspective, it would be extremely interesting to investigate the expression of DBE-T in asymmetrically affected muscles of FSHD patients, to investigate the association between the production of this lncRNA and the casual, stochastic establishment of a dystrophic phenotype only in certain axes of the body during development.
Even though methylation of H3K36 has been mainly associated with gene activation, there are reports ascribing multiple roles to this histone mark. Different biological outcomes depends on when (developmental stage or cell cycle phase) or where (promoter region or gene body) the methylated H3K36 mark is placed and on which histone reader binds to this modification.Citation66 While in budding yeast all levels of H3K36 methylation are executed by a single enzyme, termed Set2, which is coupled to transcriptional elongation,Citation67 in mammals multiple, non redundant proteins are responsible for the different flavors of H3K36 methylation, clearly suggesting an increase in function complexity during evolution. In particular, H3K36me2 has been linked to gene activation,Citation68,Citation69 prevention of spurious intragenic transcription initiationCitation70 and in the cellular response to DNA double-strand breaks.Citation71 In agreement with a recent study,Citation69 we observed enrichment of H3K36me2 in proximity of DBE-T TSS.Citation18 Notably, Ash1L knock down leads to reduced H3K36me2 accumulation and decreased target expression, supporting an activatory role for this histone mark in our system.
Intriguingly, it appears to exist an evolutionary conserved antagonism between H3K36 and H3K27 methylation.Citation72-Citation74 Considering that both the PcG hallmark H3K27me3 and H3K36me2 peak in proximity of DBE-T TSS, it would be interesting to better characterize the relationship between these two marks in the context of FSHD.
Conclusions
Transcripts arising from repeats are cell-type specific and source of regulatory information, as repeats can provide alternative promoters, alternative exons, regulatory ncRNAs and short interfering RNAs targeting cellular genes.Citation75-Citation79 The influence of repeat transcription to the transcriptional output of mammalian genomes is surprisingly broad. In mammalian cells, around 6-30% of the total amount of transcripts initiate within repetitive elements.Citation77 Hence, transcription of repetitive elements could be one of the major driving forces of evolution.
The majority of the typical PcG histone marks are located outside protein-coding regions and mainly in genomic repeats.Citation31,Citation80-Citation83 Our results support the hypothesis that repetitive elements might function as genomic binding platforms for PcG, whose activity can be regulated by lncRNAs.Citation18,Citation83
RNAs are arbitrarily defined as non-protein coding if they do not contain an ORF longer than 100 amino acids.Citation4,Citation84 Nevertheless, this dichotomy does not necessarily reflect distinct biological functions.Citation4,Citation84-Citation86 Indeed, there are examples of protein-coding RNAs displaying also non-coding functions.Citation87-Citation93 Moreover, RNAs encoding for small (less than 100 amino acids) functional peptides have been reported.Citation94 Thus, the complexity of RNA functions has just begun to be unveiled and upcoming discoveries might reveal unexpected surprises.
Acknowledgements
This work is a partial fulfillment of Valentina Casa’s PhD in Molecular Medicine, Program in Neuroscience, San Raffaele University. The Gabellini laboratory is supported by the European Research Council (ERC), the Italian Epigenomics Flagship Project, the Italian Ministry of Health and the FSHD Global Research Foundation. D. Gabellini is a Dulbecco Telethon Institute Assistant Scientist.
References
- Mattick JS. Deconstructing the dogma: a new view of the evolution and genetic programming of complex organisms. Ann N Y Acad Sci 2009; 1178:29 - 46; http://dx.doi.org/10.1111/j.1749-6632.2009.04991.x; PMID: 19845626
- Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res 2012; 40:6391 - 400; http://dx.doi.org/10.1093/nar/gks296; PMID: 22492512
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al, FANTOM Consortium, RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group). The transcriptional landscape of the mammalian genome. Science 2005; 309:1559 - 63; http://dx.doi.org/10.1126/science.1112014; PMID: 16141072
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009; 10:155 - 9; http://dx.doi.org/10.1038/nrg2521; PMID: 19188922
- Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res 2011; 90:430 - 40; http://dx.doi.org/10.1093/cvr/cvr097; PMID: 21558279
- Derrien T, Guigó R, Johnson R. The Long Non-Coding RNAs: A New (P)layer in the “Dark Matter”. Front Genet 2011; 2:107; PMID: 22303401
- Orom UA, Shiekhattar R. Noncoding RNAs and enhancers: complications of a long-distance relationship. Trends Genet 2011; 27:433 - 9; http://dx.doi.org/10.1016/j.tig.2011.06.009; PMID: 21831473
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 2011; 7:e1002384; http://dx.doi.org/10.1371/journal.pgen.1002384; PMID: 22144907
- Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol 2012; 30:99 - 104; http://dx.doi.org/10.1038/nbt.2024; PMID: 22081020
- Spitale RC, Tsai MC, Chang HY. RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics 2011; 6:539 - 43; http://dx.doi.org/10.4161/epi.6.5.15221; PMID: 21393997
- Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science 2006; 311:1118 - 23; http://dx.doi.org/10.1126/science.1117705; PMID: 16497925
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007; 129:1311 - 23; http://dx.doi.org/10.1016/j.cell.2007.05.022; PMID: 17604720
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008; 322:750 - 6; http://dx.doi.org/10.1126/science.1163045; PMID: 18974356
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA 2009; 106:11667 - 72; http://dx.doi.org/10.1073/pnas.0904715106; PMID: 19571010
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011; 472:120 - 4; http://dx.doi.org/10.1038/nature09819; PMID: 21423168
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011; 477:295 - 300; http://dx.doi.org/10.1038/nature10398; PMID: 21874018
- Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell 2011; 43:1040 - 6; http://dx.doi.org/10.1016/j.molcel.2011.08.019; PMID: 21925392
- Cabianca DS, Casa V, Bodega B, Xynos A, Ginelli E, Tanaka Y, et al. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell 2012; 149:819 - 31; http://dx.doi.org/10.1016/j.cell.2012.03.035; PMID: 22541069
- Guil S, Soler M, Portela A, Carrère J, Fonalleras E, Gómez A, et al. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat Struct Mol Biol 2012; 19:664 - 70; http://dx.doi.org/10.1038/nsmb.2315; PMID: 22659877
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 1961; 190:372 - 3; http://dx.doi.org/10.1038/190372a0; PMID: 13764598
- Lee JT. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat Rev Mol Cell Biol 2011; 12:815 - 26; http://dx.doi.org/10.1038/nrm3231; PMID: 22108600
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 2009; 10:697 - 708; PMID: 19738629
- Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol 2011; 12:799 - 814; http://dx.doi.org/10.1038/nrm3230; PMID: 22108599
- Martinez AM, Cavalli G. The role of polycomb group proteins in cell cycle regulation during development. Cell Cycle 2006; 5:1189 - 97; http://dx.doi.org/10.4161/cc.5.11.2781; PMID: 16721063
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer 2006; 6:846 - 56; http://dx.doi.org/10.1038/nrc1991; PMID: 17060944
- Lin YW, Chen HM, Fang JY. Gene silencing by the Polycomb group proteins and associations with cancer. Cancer Invest 2011; 29:187 - 95; http://dx.doi.org/10.3109/07357907.2010.512605; PMID: 21294604
- Gieni RS, Hendzel MJ. Polycomb group protein gene silencing, non-coding RNA, stem cells, and cancer. Biochem Cell Biol 2009; 87:711 - 46; http://dx.doi.org/10.1139/O09-057; PMID: 19898523
- Kanduri C, Whitehead J, Mohammad F. The long and the short of it: RNA-directed chromatin asymmetry in mammalian X-chromosome inactivation. FEBS Lett 2009; 583:857 - 64; http://dx.doi.org/10.1016/j.febslet.2009.02.004; PMID: 19302783
- Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet 2002; 30:167 - 74; http://dx.doi.org/10.1038/ng820; PMID: 11780141
- Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell 2011; 146:119 - 33; http://dx.doi.org/10.1016/j.cell.2011.06.026; PMID: 21729784
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, et al. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J 2006; 25:3110 - 22; http://dx.doi.org/10.1038/sj.emboj.7601187; PMID: 16763550
- Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet 1999; 21:400 - 4; http://dx.doi.org/10.1038/7734; PMID: 10192391
- Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell 2006; 21:617 - 28; http://dx.doi.org/10.1016/j.molcel.2006.01.028; PMID: 16507360
- Hogga I, Karch F. Transcription through the iab-7 cis-regulatory domain of the bithorax complex interferes with maintenance of Polycomb-mediated silencing. Development 2002; 129:4915 - 22; PMID: 12397100
- Rank G, Prestel M, Paro R. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol Cell Biol 2002; 22:8026 - 34; http://dx.doi.org/10.1128/MCB.22.22.8026-8034.2002; PMID: 12391168
- Schmitt S, Prestel M, Paro R. Intergenic transcription through a polycomb group response element counteracts silencing. Genes Dev 2005; 19:697 - 708; http://dx.doi.org/10.1101/gad.326205; PMID: 15741315
- Sessa L, Breiling A, Lavorgna G, Silvestri L, Casari G, Orlando V. Noncoding RNA synthesis and loss of Polycomb group repression accompanies the colinear activation of the human HOXA cluster. RNA 2007; 13:223 - 39; http://dx.doi.org/10.1261/rna.266707; PMID: 17185360
- Flanigan KM, Coffeen CM, Sexton L, Stauffer D, Brunner S, Leppert MF. Genetic characterization of a large, historically significant Utah kindred with facioscapulohumeral dystrophy. Neuromuscul Disord 2001; 11:525 - 9; http://dx.doi.org/10.1016/S0960-8966(01)00201-2; PMID: 11525880
- Cabianca DS, Gabellini D. The cell biology of disease: FSHD: copy number variations on the theme of muscular dystrophy. J Cell Biol 2010; 191:1049 - 60; http://dx.doi.org/10.1083/jcb.201007028; PMID: 21149563
- Hewitt JE, Lyle R, Clark LN, Valleley EM, Wright TJ, Wijmenga C, et al. Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum Mol Genet 1994; 3:1287 - 95; http://dx.doi.org/10.1093/hmg/3.8.1287; PMID: 7987304
- Winokur ST, Bengtsson U, Feddersen J, Mathews KD, Weiffenbach B, Bailey H, et al. The DNA rearrangement associated with facioscapulohumeral muscular dystrophy involves a heterochromatin-associated repetitive element: implications for a role of chromatin structure in the pathogenesis of the disease. Chromosome Res 1994; 2:225 - 34; http://dx.doi.org/10.1007/BF01553323; PMID: 8069466
- Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, Dauwerse HG, et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet 1992; 2:26 - 30; http://dx.doi.org/10.1038/ng0992-26; PMID: 1363881
- van Deutekom JC, Wijmenga C, van Tienhoven EA, Gruter AM, Hewitt JE, Padberg GW, et al. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet 1993; 2:2037 - 42; http://dx.doi.org/10.1093/hmg/2.12.2037; PMID: 8111371
- Chadwick BP. Macrosatellite epigenetics: the two faces of DXZ4 and D4Z4. Chromosoma 2009; 118:675 - 81; http://dx.doi.org/10.1007/s00412-009-0233-5; PMID: 19690880
- Giacalone J, Friedes J, Francke U. A novel GC-rich human macrosatellite VNTR in Xq24 is differentially methylated on active and inactive X chromosomes. Nat Genet 1992; 1:137 - 43; http://dx.doi.org/10.1038/ng0592-137; PMID: 1302007
- Kogi M, Fukushige S, Lefevre C, Hadano S, Ikeda JE. A novel tandem repeat sequence located on human chromosome 4p: isolation and characterization. Genomics 1997; 42:278 - 83; http://dx.doi.org/10.1006/geno.1997.4746; PMID: 9192848
- Neguembor MV, Gabellini D. In junk we trust: repetitive DNA, epigenetics and facioscapulohumeral muscular dystrophy. Epigenomics 2010; 2:271 - 87; http://dx.doi.org/10.2217/epi.10.8; PMID: 22121874
- van Overveld PG, Lemmers RJ, Sandkuijl LA, Enthoven L, Winokur ST, Bakels F, et al. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet 2003; 35:315 - 7; http://dx.doi.org/10.1038/ng1262; PMID: 14634647
- Bodega B, Ramirez GD, Grasser F, Cheli S, Brunelli S, Mora M, et al. Remodeling of the chromatin structure of the facioscapulohumeral muscular dystrophy (FSHD) locus and upregulation of FSHD-related gene 1 (FRG1) expression during human myogenic differentiation. BMC Biol 2009; 7:41; http://dx.doi.org/10.1186/1741-7007-7-41; PMID: 19607661
- Zeng W, de Greef JC, Chen YY, Chien R, Kong X, Gregson HC, et al. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD). PLoS Genet 2009; 5:e1000559; http://dx.doi.org/10.1371/journal.pgen.1000559; PMID: 19593370
- Petrov A, Pirozhkova I, Carnac G, Laoudj D, Lipinski M, Vassetzky YS. Chromatin loop domain organization within the 4q35 locus in facioscapulohumeral dystrophy patients versus normal human myoblasts. Proc Natl Acad Sci USA 2006; 103:6982 - 7; http://dx.doi.org/10.1073/pnas.0511235103; PMID: 16632607
- Pirozhkova I, Petrov A, Dmitriev P, Laoudj D, Lipinski M, Vassetzky Y. A functional role for 4qA/B in the structural rearrangement of the 4q35 region and in the regulation of FRG1 and ANT1 in facioscapulohumeral dystrophy. PLoS ONE 2008; 3:e3389; http://dx.doi.org/10.1371/journal.pone.0003389; PMID: 18852887
- Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell 2002; 110:339 - 48; http://dx.doi.org/10.1016/S0092-8674(02)00826-7; PMID: 12176321
- Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camaño P, Dauwerse JG, et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 2010; 329:1650 - 3; http://dx.doi.org/10.1126/science.1189044; PMID: 20724583
- Lemmers RJ, Osborn M, Haaf T, Rogers M, Frants RR, Padberg GW, et al. D4F104S1 deletion in facioscapulohumeral muscular dystrophy: phenotype, size, and detection. Neurology 2003; 61:178 - 83; http://dx.doi.org/10.1212/01.WNL.0000078889.51444.81; PMID: 12874395
- Balog J, Thijssen PE, de Greef JC, Shah B, van Engelen BG, Yokomori K, et al. Correlation analysis of clinical parameters with epigenetic modifications in the DUX4 promoter in FSHD. Epigenetics 2012; 7:579 - 84; http://dx.doi.org/10.4161/epi.20001; PMID: 22522912
- Duncan IW. Transvection effects in Drosophila. Annu Rev Genet 2002; 36:521 - 56; http://dx.doi.org/10.1146/annurev.genet.36.060402.100441; PMID: 12429702
- Stout K, van der Maarel S, Frants RR, Padberg GW, Ropers HH, Haaf T. Somatic pairing between subtelomeric chromosome regions: implications for human genetic disease?. Chromosome Res 1999; 7:323 - 9; http://dx.doi.org/10.1023/A:1009287111661; PMID: 10515207
- Goto K, Song MD, Lee JH, Arahata K. [Genetic analysis of facioscapulohumeral muscular dystrophy (FSHD)]. Rinsho Shinkeigaku 1995; 35:1416 - 8; PMID: 8752415
- Lunt PW, Jardine PE, Koch MC, Maynard J, Osborn M, Williams M, et al. Correlation between fragment size at D4F104S1 and age at onset or at wheelchair use, with a possible generational effect, accounts for much phenotypic variation in 4q35-facioscapulohumeral muscular dystrophy (FSHD). Hum Mol Genet 1995; 4:951 - 8; http://dx.doi.org/10.1093/hmg/4.5.951; PMID: 7633457
- Zatz M, Marie SK, Passos-Bueno MR, Vainzof M, Campiotto S, Cerqueira A, et al. High proportion of new mutations and possible anticipation in Brazilian facioscapulohumeral muscular dystrophy families. Am J Hum Genet 1995; 56:99 - 105; PMID: 7825608
- Tawil R, Forrester J, Griggs RC, Mendell J, Kissel J, McDermott M, et al, The FSH-DY Group. Evidence for anticipation and association of deletion size with severity in facioscapulohumeral muscular dystrophy. Ann Neurol 1996; 39:744 - 8; http://dx.doi.org/10.1002/ana.410390610; PMID: 8651646
- Hsu YD, Kao MC, Shyu WC, Lin JC, Huang NE, Sun HF, et al. Application of chromosome 4q35-qter marker (pFR-1) for DNA rearrangement of facioscapulohumeral muscular dystrophy patients in Taiwan. J Neurol Sci 1997; 149:73 - 9; http://dx.doi.org/10.1016/S0022-510X(97)05394-X; PMID: 9168169
- Ricci E, Galluzzi G, Deidda G, Cacurri S, Colantoni L, Merico B, et al. Progress in the molecular diagnosis of facioscapulohumeral muscular dystrophy and correlation between the number of KpnI repeats at the 4q35 locus and clinical phenotype. Ann Neurol 1999; 45:751 - 7; http://dx.doi.org/10.1002/1531-8249(199906)45:6<751::AID-ANA9>3.0.CO;2-M; PMID: 10360767
- Bumgarner SL, Neuert G, Voight BF, Symbor-Nagrabska A, Grisafi P, van Oudenaarden A, et al. Single-cell analysis reveals that noncoding RNAs contribute to clonal heterogeneity by modulating transcription factor recruitment. Mol Cell 2012; 45:470 - 82; http://dx.doi.org/10.1016/j.molcel.2011.11.029; PMID: 22264825
- Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol 2012; 13:115 - 26; http://dx.doi.org/10.1038/nrm3274; PMID: 22266761
- Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol 2005; 25:3305 - 16; http://dx.doi.org/10.1128/MCB.25.8.3305-3316.2005; PMID: 15798214
- Rao B, Shibata Y, Strahl BD, Lieb JD. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol Cell Biol 2005; 25:9447 - 59; http://dx.doi.org/10.1128/MCB.25.21.9447-9459.2005; PMID: 16227595
- Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J, et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell 2011; 44:609 - 20; http://dx.doi.org/10.1016/j.molcel.2011.08.042; PMID: 22099308
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 2005; 123:581 - 92; http://dx.doi.org/10.1016/j.cell.2005.10.023; PMID: 16286007
- Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M, et al. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci USA 2011; 108:540 - 5; http://dx.doi.org/10.1073/pnas.1013571108; PMID: 21187428
- Bender LB, Suh J, Carroll CR, Fong Y, Fingerman IM, Briggs SD, et al. MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development 2006; 133:3907 - 17; http://dx.doi.org/10.1242/dev.02584; PMID: 16968818
- Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol 2007; 9:804 - 12; http://dx.doi.org/10.1038/ncb1608; PMID: 17589499
- Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem 2011; 286:7983 - 9; http://dx.doi.org/10.1074/jbc.M110.194027; PMID: 21239496
- Norris J, Fan D, Aleman C, Marks JR, Futreal PA, Wiseman RW, et al. Identification of a new subclass of Alu DNA repeats which can function as estrogen receptor-dependent transcriptional enhancers. J Biol Chem 1995; 270:22777 - 82; http://dx.doi.org/10.1074/jbc.270.39.22777; PMID: 7559405
- Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol 2001; 21:1973 - 85; http://dx.doi.org/10.1128/MCB.21.6.1973-1985.2001; PMID: 11238933
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 2009; 41:563 - 71; http://dx.doi.org/10.1038/ng.368; PMID: 19377475
- Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 2011; 471:325 - 30; http://dx.doi.org/10.1038/nature09830; PMID: 21297615
- Shen S, Lin L, Cai JJ, Jiang P, Kenkel EJ, Stroik MR, et al. Widespread establishment and regulatory impact of Alu exons in human genes. Proc Natl Acad Sci USA 2011; 108:2837 - 42; http://dx.doi.org/10.1073/pnas.1012834108; PMID: 21282640
- Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AA, Perez-Burgos L, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 2003; 12:1577 - 89; http://dx.doi.org/10.1016/S1097-2765(03)00477-5; PMID: 14690609
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 2004; 7:663 - 76; http://dx.doi.org/10.1016/j.devcel.2004.10.005; PMID: 15525528
- Day DS, Luquette LJ, Park PJ, Kharchenko PV. Estimating enrichment of repetitive elements from high-throughput sequence data. Genome Biol 2010; 11:R69; http://dx.doi.org/10.1186/gb-2010-11-6-r69; PMID: 20584328
- Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, Wutz A. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev 2010; 24:265 - 76; http://dx.doi.org/10.1101/gad.544410; PMID: 20123906
- Costa FF. Non-coding RNAs: Meet thy masters. Bioessays 2010; 32:599 - 608; http://dx.doi.org/10.1002/bies.200900112; PMID: 20544733
- Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet 2009; 5:e1000459; http://dx.doi.org/10.1371/journal.pgen.1000459; PMID: 19390609
- Martin L, Chang HY. Uncovering the role of genomic “dark matter” in human disease. J Clin Invest 2012; 122:1589 - 95; http://dx.doi.org/10.1172/JCI60020; PMID: 22546862
- Rastinejad F, Blau HM. Genetic complementation reveals a novel regulatory role for 3′ untranslated regions in growth and differentiation. Cell 1993; 72:903 - 17; http://dx.doi.org/10.1016/0092-8674(93)90579-F; PMID: 8384533
- Jupe ER, Liu XT, Kiehlbauch JL, McClung JK, Dell’Orco RT. Prohibitin in breast cancer cell lines: loss of antiproliferative activity is linked to 3′ untranslated region mutations. Cell Growth Differ 1996; 7:871 - 8; PMID: 8809404
- Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J 1998; 17:3448 - 60; http://dx.doi.org/10.1093/emboj/17.12.3448; PMID: 9628880
- Amack JD, Paguio AP, Mahadevan MS. Cis and trans effects of the myotonic dystrophy (DM) mutation in a cell culture model. Hum Mol Genet 1999; 8:1975 - 84; http://dx.doi.org/10.1093/hmg/8.11.1975; PMID: 10484765
- Jenny A, Hachet O, Závorszky P, Cyrklaff A, Weston MD, Johnston DS, et al. A translation-independent role of oskar RNA in early Drosophila oogenesis. Development 2006; 133:2827 - 33; http://dx.doi.org/10.1242/dev.02456; PMID: 16835436
- Candeias MM, Malbert-Colas L, Powell DJ, Daskalogianni C, Maslon MM, Naski N, et al. P53 mRNA controls p53 activity by managing Mdm2 functions. Nat Cell Biol 2008; 10:1098 - 105; http://dx.doi.org/10.1038/ncb1770; PMID: 19160491
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010; 465:1033 - 8; http://dx.doi.org/10.1038/nature09144; PMID: 20577206
- Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, et al. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 2010; 329:336 - 9; http://dx.doi.org/10.1126/science.1188158; PMID: 20647469