Abstract
Selenoprotein P (Sepp1), a glycoprotein rich in selenium, is thought to function in selenium transport throughout the body. The sepp1 gene locus potentially produces three alternative transcripts that differ only in their 5′ untranslated regions (5′UTRs) and not in their protein coding regions, as indicated by transcript information in genomic databases. Here we investigated the distribution, relative expression, and biological significance of these transcript variants. We confirmed the expression of Sepp1 transcript variants using PCR and sequencing. Using 5′-RACE, we identified multiple 5′-termini upstream from three different splice donor sites, and a single splice acceptor site for exon 2. We found regional and temporal changes in variant expression in select adult and neonate murine tissue and brain regions. Distribution of variants in heart and kidney varied with stage of development. Notably, the Sepp1b variant was localized specifically to the hippocampus in brain. Targeted silencing of individual variants using RNAi demonstrated the biological importance for all transcript variants in cell viability. Additionally, we determined that the Sepp1b variant is a specific target for the miR-7 microRNA by means of its unique 5′UTR structure. Our results emphasize the importance of non-coding transcript variations as a regulatory means for Sepp1 expression in different tissues and stages of development. The presence of a variant localized in the hippocampus and regulated by a microRNA may have implications for the known deficits in synaptic function caused by genetic deletion of Sepp1.
Introduction
Recent studies have highlighted the importance of non-coding sequences in regulation of gene expression. Three murine Selenoprotein P (Sepp1) transcripts present in the National Center for Biotechnology Information (NCBI, www.ncbi.nlm.nih.gov/) nucleotide database have identical coding exons, but differ in their 5′ noncoding exons. Thus, the variants are predicted to have the same protein products, suggesting a purely regulatory role for the variants. Sepp1 transcripts are present in most mammalian tissues, with the highest expression in liver. Sepp1 is secreted primarily from liver cells and acts as a carrier to bring selenium (Se) to brain and other body regions.Citation1 In addition, Sepp1 has antioxidant propertiesCitation2,Citation3 and can chelate toxic metals.Citation4,Citation5 Sepp1 knockout (KO) animals are deficient in synaptic plasticity in the hippocampus, suggesting an important role for Sepp1 other than selenium transport.Citation6
Selenoproteins contain the 21st amino acid selenocysteine (Sec), a structural analog of cysteine with Se in place of sulfur.Citation7 The codon for Sec is UGA, which is a stop codon for non-selenoprotein mRNAs. The selenoprotein family requires the non-coding selenocysteine insertion sequence (SECIS) element in the 3′ UTR of the mRNAs for translation.Citation8 The SECIS element binds factors necessary for the recoding of UGA from a stop codon to the codon for Sec.Citation7 Sec is synthesized directly on its own tRNA (tRNASEC) through charging of the tRNA with a serine residue, followed by phosphorylation of the serine and subsequent exchange with selenophosphate. Sepp1 is unusual in having multiple Sec residues (10 in rodents and primates),Citation9 as well as two SECIS elements in the 3′ UTR.Citation10
MicroRNAs (miRNAs) are a large class of endogenous non-coding RNAs that are typically 21–22 nucleotides in length.Citation11 miRs can suppress translation of mRNAs or target them for degradation.Citation12 High complementarity of the miRNA target in mRNAs to a “seed” region in the miRNA in nucleotides 2–8 is important for miRNA target recognition.Citation13 Endogenous miRNA target sites have primarily been found in the 3′ UTRs of their target mRNAs. This may be due in part to the commonly used target prediction algorithms, which take into account the conservation of sequences across species. Only 3′ UTRs are used for these algorithms, as conservation in coding sequences is more likely due to protein function.Citation14 5′ UTR miRNA target sites can also result in translational repression.Citation15 The hepatitis C virus utilizes the liver-specific miR-122 for positive translational regulation via a 5′ UTR target.Citation16 miR-10a binds to the 5′ UTR of ribosomal protein mRNAs, enhancing translation.Citation17 A recently discovered class of miRNAs requires simultaneous binding to 5′ and 3′ UTRs.Citation18 Thus miRNAs can target both 5′ and 3′ UTRs and exert bidirectional regulation of protein expression. We have identified a target site for the murine homolog of miR-7 in the 5′ UTR of one of the Sepp1 transcript variants.
In this study, we investigated the tissue and developmental expression of these variants, the effects of targeted knockdown of expression of individual transcripts on cell viability, and the role of miR-7 in regulation of Sepp1 expression.
Results
Genomics studies have identified three confirmed variants for mouse Sepp1, and these variants appear in the NCBI gene database. The variants all have identical coding exons (exons 2–5), but differ in their 5′ noncoding exons (exons 1a, b, and c). As the different first exons are non-coding, the variants are all predicted to have the same protein product, suggesting a purely regulatory role for the variants. This finding provides a possible mechanism for differential regulation of expression of Sepp1 in response to various stimuli, possibly including damage or disease states. and B show the arrangement and sequence information for the mouse exons within the genomic DNA. Exon 1a is closest to exon 2 on the genomic sequence, while exon 1b is the furthest 5′ exon. Exon 1c begins 44 bases 3′ of the start of exon 1b, and overlaps 1b by 30 bases.
Figure 1. Arrangement of alternative 1st exons in genomic DNA sequences. (A) Schematic showing the arrangement of mouse Sepp1 exons 1a, 1b, and 1c within the genomic DNA sequence. (B) Genomic sequence of alternative 1st exons. Exon 1a is shown in green, exon 1b is shown in red, and exon 1c is shown in blue, with purple showing the overlap region between exon 1b and exon 1c. Triangles indicate transcription start sites according to NCBI. (C) Alignment of mouse, rat and human genomic sequences showing the relative location of the alternative 5′ exons.
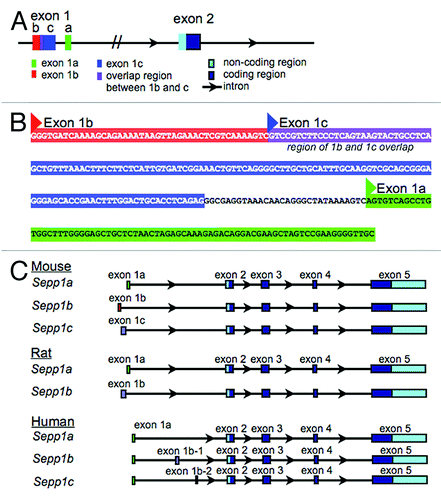
There are also three human and two rat transcript variants in NCBI and other public databases. shows the alignment of alternative 5′ exons for mouse, rat and human Sepp1. As with the mouse variants, the rat transcript variants have alternate 5′ non-coding exons. However, all sequences of all human variants begin with the same non-coding exon. Two of the human variants have an additional exon inserted after the first exon. The 5′ exon for the mouse Sepp1a variant has 81% sequence homology with the 5′ exon of the rat Sepp1a variant, and 60% homology with the human 5′ exon. Additionally, the first exon of the mouse Sepp1c variant has 80% homology with the first exon of the rat Sepp1b variant.
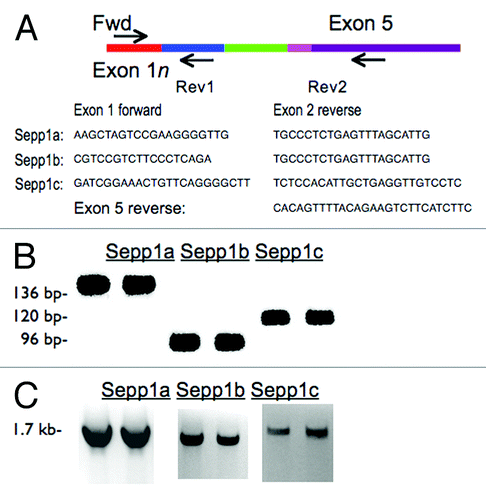
To verify these sequences, we designed PCR primer pairs with forward primers specific to each of the variant exons and reverse primers in exon 2 for verification with reverse transcriptase PCR (RT-PCR). Resulting products were verified by size and by subsequent sequencing. To determine if the PCR products were part of full-length messages, we also performed RT-PCR with exon 1 forward primers and reverse primers in exon 5 (). These products were also confirmed by size and sequencing to be the predicted variants.
Figure 2. Confirmation of transcript variants with RT-PCR. (A) cDNA map of Sepp1 showing the locations of forward and reverse primers used. Primer sequences are listed below. (B) RT-PCR products using primers in exon 1a, 1b, or 1c and reverse primers in exon 2 (as indicated by “Rev1” in cDNA map above) giving Sepp1a, Sepp1b, and Sepp1c products, respectively. (C) RT-PCR products for various tissues using the same forward primers as A, but with the reverse primers in the last exon, exon 5, as indicated by “Rev2” in map above.
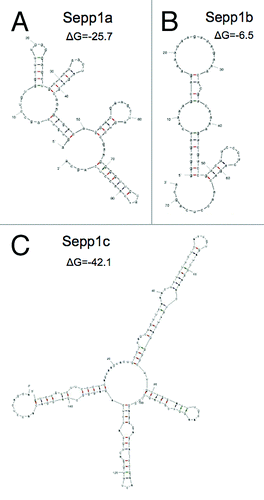
We speculated that the variants could have different rates of translation if their secondary structures were different. We therefore used the mfold software (http://mfold.rna.albany.edu) to examine the secondary structures of the variants. Resulting folds and mRNA free energies are shown in . Sepp1a is predicted to exhibit a moderate amount of secondary structure, with a Gibb’s free energy value (ΔG) of -25.7. In contrast, Sepp1b had a simpler predicted structure with a ΔG of only -6.5. Sepp1c had the most complex predicted structure, with a ΔG of -42.1. The great differences in complexity and free energy of the variants could serve to exert control over the rate and efficiency of translation of Sepp1.
Figure 3. Predicted secondary structures of 5′ transcript variants of mSepp1 and relative folding energies. (A) Transcript Sepp1a. (B) Sepp1b. C: Sepp1c.
We subsequently sought to accurately determine the 5′ termini for each variant using Rapid Amplification of 5′ DNA Ends (5′-RACE). Resulting sequences are shown in . Underlined regions correspond to the highlighted genomic sequences are shown in , with the first letter of the sequence in bold. Several possible 5′-terminal sequences were found for each transcript variant that matched the genomic DNA sequence. However, all variants had one of three splice donor sites matching those of the three predicted variants, and only one splice acceptor site for exon 2 was identified. Longer sequences are more likely to represent the true 5′ ends, although shorter exon sequences could also result from alternative transcription start sites. Most remarkable was the discovery of a sequence for the Sepp1a variant extending 84 nucleotides beyond the 5′ terminus found in the public database. This variant extends into the Sepp1c variant region on the genomic DNA. We did find several sequences corresponding to the predicted 5′ end for this variant (second sequence shown for Sepp1a), as well as several forms differing in length by 1–3 nucleotides. Interestingly, Sepp1a isolated from adult female brain was shorter by 1–3 nucleotides on the 5′ end compared with adult male brain, implying gender-specific transcription start sites. We also found a sequence for Sepp1b that extended 12 nucleotides longer in the 5′ direction than the database sequence for this variant. Two sequences for Sepp1c were shorter than found in databases; however, we cannot be certain we found the full-length 5′ sequence for this variant.
Table 1. Determination of 5′-termini using 5′-RACE
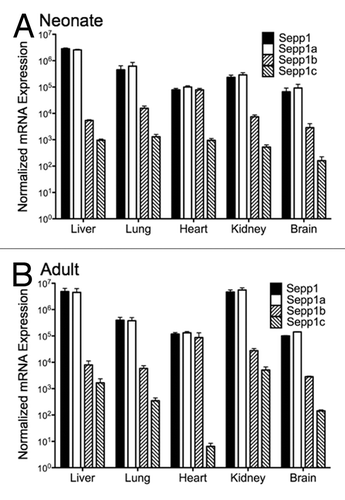
PCR primers for each of the 5′ exons shown in were then used for quantitative RT-PCR (qPCR) to measure their relative abundance in neonatal and adult mouse tissues. This revealed tissue and developmental stage specific expression patterns in neonatal and adult brain, liver, heart, and kidney (). The most abundant transcript variant in all tissues was Sepp1a. However, all variants showed temporal and tissue-specific differences. In both neonatal and adult animals, Sepp1b expression relative to total Sepp1 expression was much higher in heart than in other tissues. The Sepp1c variant in heart decreased in relative expression from neonatal animals to adult animals. One surprising difference was that kidney mRNA levels for total Sepp1 as well as the Sepp1a variant were higher in adult animals than in neonatal animals, and were comparable in expression in liver and kidney. Thus while Sepp1 synthesis in liver is important for Se delivery, kidney may also synthesize Sepp1, perhaps for recycling whole body Se.
Figure 4. Expression of Sepp1 transcript variants in different organs for total Sepp1, Sepp1a, Sepp1 b, and Sepp1c from neonatal (A) and adult (B) animals. Absolute measurements of qPCR from indicated tissue were performed using primers specific for each Sepp1 transcript variant, calculated using the primer efficiencies for that variant, and normalized to product from 18S rRNA primers.
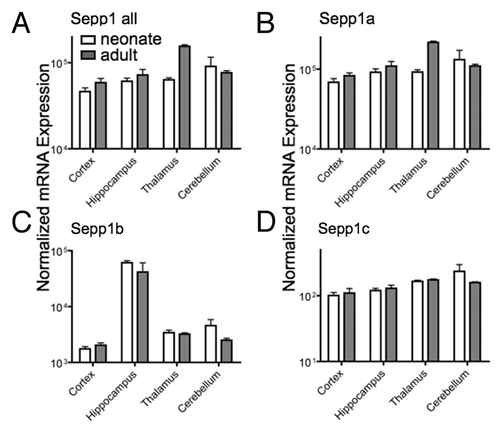
We next assessed changes during development in different brain regions. Although Sepp1a was the predominant variant in brain, we found specific regional and development expression patterns for all three variants (). Notably, the Sepp1b transcript expression was greater in the hippocampus compared with other brain regions, and may be have important for hippocampal functions that are impaired in Sepp1 KO animals. Additionally, total Sepp1 and the Sepp1a variant increase in thalamus from neonates to adults. These data show a high degree of spatial and temporal regulation of Sepp1 transcript variants.
Figure 5. Expression of Sepp1 transcript variants in different brain regions for total Sepp1 (A), Sepp1a (B), Sepp1 b(C), and Sepp1c (D). Absolute measurements of qPCR from indicated tissue were performed as described in .
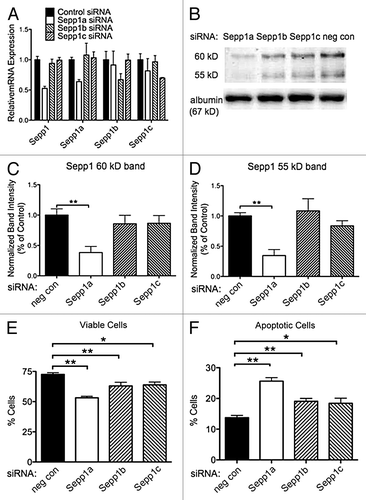
We previously reported that knocking down Sepp1 expression with shRNA decreases viability of cultured N2A cells.Citation19 To determine which variants of Sepp1 may be necessary for viability of these cells, we used RNAi directed toward the three Sepp1 transcripts. Cultured N2A cells were transfected with siRNA specific to each variant. Specificity of knockdown was demonstrated by qPCR (). We found that siRNA directed to the Sepp1a variant resulted in a significant decrease in the 60 kD band of Sepp1 (). Knockdown with siRNAs targeting Sepp1b and Sepp1c produced a smaller decrease in Sepp1 expression that was not significant. Sepp1a siRNA also significantly reduced expression of the 55 kD Sepp1 band (), with a smaller reduction from Sepp1c siRNA and notably no decrease from Sepp1b siRNA. Knockdown of Sepp1a produced the greatest decrease in viable cells (), and the greatest increase in apoptotic cells (, Annexin V positive). However, knockdown of each transcript resulted in decreased viability and increased apoptotic cells, compared with the non-targeting siRNA knockdown. This suggests that all three transcripts may be important for neuronal cell viability in different ways.
Figure 6. Knockdown of Sepp1 transcript variants with siRNA increases apoptosis after oxidative challenge. Viability and apoptosis were assessed by flow cytometry for propidium iodide and Annexin V fluorescence following RNAi-mediated knockdown of Sepp1a, b or c transcripts in N2a murine neuronal cells. (A) Confirmation of siRNA efficacy and specificity by qPCR measurement of RNA normalized to 18S rRNA for total Sepp1 and for each variant, shown as median ± range. (B) western blot (above) showing Sepp1 in media of N2A cells following siRNA transfection, and the 67 kD albumin band (below). The (C) Quantification of the 60 kD Sepp1 band using albumin as a loading control, relative to negative control siRNA samples. (D) Quantification of 55 kD band. (E) Percent viable cells negative for both propidium iodine and annexin V. (F) Percent apoptotic cells defined as negative for propidium iodide but positive for annexin V. * indicates p < 0.05 and ** indicates p < 0.01, 2-way ANOVA with Bonferonni’s posthoc test.
To determine how the non-coding alternative exons could regulate expression of Sepp1, we used various search algorithms to look for regulatory elements in the 5′ UTR. These included UTRscan (http://itbtools.ba.itb.cnr.it/utrscan) and RegRNA (regrna.mbc.nctu.edu.tw/html/prediction.html). One intriguing element found with both searches was a “GY-box” for both the Sepp1b and Sepp1c variants, in the region where they overlap on the genomic DNA. This sequence was first found as a repressive element in the 3′ UTR of genes in the Notch pathway involved in neural development of Drosophila,Citation20,Citation21 and was later found to be a target site for the miR-7 microRNA.Citation22 We examined the transcript variant sequences to determine if they could provide target sites for this miRNA. Although most miRNAs target 3′ UTRs, functional miRNA sites have been demonstrated in 5′UTRs as well.Citation17
As shown below, the murine homolog of miR-7 aligns with a high degree of complementarity to the Sepp1b transcript variant. The GY-box (shown in bold on the Sepp1b sequence) provides a seven nucleotide “seed” region on bases 2–7 of miR-7 reading 5′-3′. Eleven of the following 15 bases of miR-7 pair with the Sepp1b sequence, plus 3 wobble parings, gives a total of 18 paired bases out of 22 total bases. The Sepp1a variant lacked a GY box or a miR-7 binding site. Although the Sepp1c variant has a GY box, it is close to the 5′ terminus and is unlikely to serve as the seed region for miR-7 as half of the miRNA would extend beyond the Sepp1c 5′ terminus.
To confirm this targeting, we submitted these sequences to the online search algorithm MiRTIF (MicroRNA:Target Interaction Filter, mirtif.bii.a-star.edu.sg/), which serves as a post-processing filter for predicted miRNA:target duplexes. Pairings rated with positive scores using this algorithm indicate predicted true interactions based on “training” with experimentally confirmed miRNA:target pairs. The software verified Sepp1b 5′ UTR as a candidate target for miR-7. Additionally, we confirmed the Sepp1b 5′ UTR as a possible target of miR-7 using the RNA hybrid algorithm (bibiserv.techfak.uni-bielefeld.de/rnahybrid/).
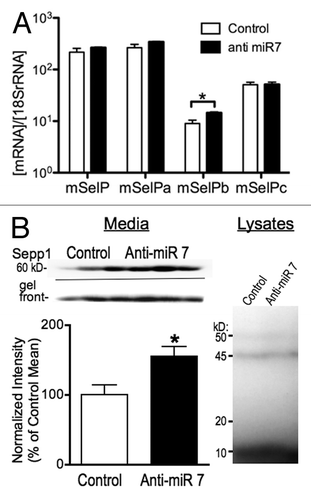
To test the possible role of miR-7 in regulating expression of Sepp1, we targeted miR-7 using a locked nucleic acid (LNA) antisense probe (Exiqon). We transfected N2A cells with an anti-miR-7 LNA or control 48 h before harvesting RNA. Subsequent qPCR demonstrated an increase in mSepp1b mRNA (). Since miRNAs can repress gene regulation without causing RNA degradation,Citation23 we examined protein expression by labeling selenoproteins with 75Se. Media from transfected cells primarily showed one band of about 60 kD. A second band approximately 50 kD in size was detectable but was less than one tenth the intensity of the larger band, indicating it was the N-terminal region with only one Sec residue. It was not practical to quantitate this band because of the low relative intensity. LNA knockdown of miR-7 increased the expression of the 60 kD band of Sepp1 by more than 50% (, left). In contrast, we found no difference in expression of other selenoproteins within the cell lysate (, right), indicating that miR-7 does not indirectly alter Sepp1 expression through targeting of other factors controlling selenoprotein synthesis or other off target mechanisms. Thus miR-7 may regulate expression of Sepp1, possibly by repressing translation of the Sepp1b transcript variant.
Figure 7. Knockdown of miR-7 with an antisense LNA probe increases expression of Sepp1. (A) qPCR measurement of mRNA expression for Sepp1 and transcript variants 48 h following transfection of N2A cells with LNA antisense probe, normalized to 18S rRNA. (B) Protein expression measured by autoradiography of 75Se-labeled protein from N2A cell cultures 72 h after knockdown of miR-7. Above: Autoradiography of media from three control (left) and three anti-miR-7 transfected (right) cultures. Below left: Graph of 75Se-Sepp1 expression normalized to relative radioactivity for each sample. * indicates p < 0.05, two-tailed Student’s T-test. Below right: Autoradiography of N2A cell lysates.
Discussion
In this study, we demonstrated temporal tissue-specific differences in expression of three predicted Sepp1 transcript variants, and showed that knocking down all variants in a neuronal cell line decreased cell viability. Importantly, we demonstrate a novel mechanism for gene regulation that utilizes miRNA targeting of 5′ alternative exons. We found several possible 5′ end sequences of these variants, but only three potential donor sites on the 3′ end of the 1st exon sequences, all sharing a common acceptor site for a single exon 2.
Although the different transcription start sites would result in many variants, we can categorize these variants into three groups based on the three possible splice donor sites on the 3′ end of these exons. The groups correspond to the three variants identified in NCBI and other databases. While it is possible that some sequences we identified were somehow truncated by experimental procedures or even by cellular processes prior to isolation, it appears that different 5′ ends for each variant group do exist, most likely as the result of variations in transcription start sites. In addition, we identified sequences for the variant a and b groups that extend further 5′ than the sequences listed in NCBI. Interestingly, we found no overlap for the 5′ ends for each variant group between RNAs isolated from male vs female brain. Previous gender differences in selenoprotein expression have been described,Citation24 and the severity of behavioral impairments resulting from deletion of Sepp1 in mice depends on gender.Citation25 Sepp1 was previously isolated as an androgen-responsive gene,Citation26 indicating gender-specific gene regulation. Additionally, gender differences in human Sepp1 single nucleotide polymorphisms (SNPs) have previously been identified.Citation27 Thus different promoter elements may regulate Sepp1 expression in males and females, resulting in different transcription start sites.
The finding of a non-coding transcript variant that is primarily expressed in hippocampus is of particular interest. Sepp1 KO mice are deficient in hippocampal long-term potentiation (LTP), a cellular model for learning and memory.Citation28 Our finding suggests the possible involvement of Sepp1 produced in hippocampus in LTP. Sepp1 binds to apolipoprotein E receptor 2 (ApoER2).Citation29,Citation30 Another brain ligand for ApoER2 is the brain signaling protein reelin, which can enhance LTP.Citation31,Citation32 The LTP deficit in Sepp1 KO animals indicates that Sepp1 may have a similar influence on LTP. The presence of a specific Sepp1 variant suggests a local signaling role for Sepp1 in the hippocampus. Regulation of the variant by microRNA could therefore greatly influence learning and memory. Interestingly, miR-7 in humans is decreased in glioblastoma,Citation33,Citation34 and also regulates the Parkinson disease-related protein α-synuclein.Citation35 Importantly, miR-7 also increases with differentiation and development of neurons, and controls neurite outgrowth.Citation36 It should be noted that a previous study by Zhang et al. (2008),Citation37 based on analysis of expression in the Allan Brain Atlas (http://www.brain-map.org/), reported that Sepp1 expression was lower in hippocampus relative to other brain regions. However, the data set available at the time of that study is now reported to have failed quality control (http://mouse.brain-map.org/experiment/show/68076896). A newer data set (http://mouse.brain-map.org/experiment/show/79903768) shows much higher expression in hippocampus. Our findings of the relative expression of Sepp1 in adult brain, being (highest to lowest) thalamus, cerebellum, hippocampus and cortex, are consistent with the online data. Additionally, a previous study using single-cell microarray analysis of mouse brain neurons reported Sepp1 as having relatively high expression in GABAergic neurons of the hippocampus.Citation38 Thus Sepp1 appears to be synthesized in neurons and be important for brain function.
The developmental increase in Sepp1 transcript expression in kidney was surprising, as Sepp1 is thought to primarily be synthesized in liver for delivery of Se to the brain, testes and other organs.Citation39 Although Sepp1 binds to the ApoER2 receptor in brain and testes,Citation29,Citation30 binding to the megalin receptor in kidney is important to prevent urinary excretion of Sepp1.Citation40,Citation41 Higher expression of Sepp1 following development could indicate recycling of bodily Se. The reason for synthesizing new Sepp1 in kidney is unclear, but possibly the kidney acts as a filter for oxidized or otherwise modified forms of Sepp1.
Splice variants differing only in 5′UTRs have been demonstrated for several genes, including the glucocorticoid receptor,Citation42 adenosine A2A receptor,Citation43 and thyroid hormone receptor α (TRα).Citation44 These 5′ variants all exert regulatory control over protein translation. Efficiency of translation can be affected by Gibb’s free energy of the 5′ UTR and relative structure complexity, by the presence of elements such as the iron response element,Citation45 and also by short open reading frames (ORFs) upstream of the actual start codon.Citation46 The different 5′UTRs do not contain any upstream ORFs. The mSepp1 variants vary greatly in relative ΔG values, suggesting the complexity of folding for the alternate 5′ UTRs alone can influence the efficiency of protein translation. Sepp1 depends on control of translation efficiency to prevent premature termination of translation at the UGA Sec codon.Citation10 We have not identified other known RNA regulatory elements in the 5′UTRs, with the exception of the GY-box/miR-7 binding site.
MicroRNAs have been identified in high numbers in the nervous system, and are expressed with temporal or spatial specificity, suggesting crucial functions in regulation of their target mRNAs. miRNAs are postulated to function in modulating local translation at synapses to regulate synaptic function. According to this hypothesis, a set of miRNAs inhibits translation of target mRNAs under basal conditions, but with synaptic activity, a set of mRNAs is de-repressed to synthesize proteins capable of self-assembly into postsynaptic elements.Citation47 The presence of a miRNA site within the 5′UTR of a variant that is localized to the hippocampus within brain suggests this may be an important method of translational suppression during transport of the mRNA.
These studies provide a complex picture of translational control of mSepp1. 5′ variants with different mRNA complexity, and regional and temporal regulation could combine to tightly regulate Sepp1 expression. This regulation may be important for a protein containing a relatively high amount of the trace element selenium. The identification of a Sepp1 transcript variant enriched in hippocampus and its regulation by miRNA has implications for a role of Sepp1 in synaptic plasticity. These findings help elucidate the intricate regulation of protein synthesis by non-coding RNA and to clarify the functions of the transcript variants of this unique selenoprotein.
Materials and Methods
PCR and qPCR
Oligonucleotide primers specific to total mouse Sepp1, Sepp1a, Sepp1b, Sepp1c and 18SrRNA were designed and purchased from IDT (Coralville, IA). Total Sepp1 forward: 5′-TGT TGA AGA AGC CAT TAA GAT CG-3′; total Sepp1 reverse: 5′-CAC AGT TTT ACA GAA GTC TTC ATC TTC-3′. Sepp1a forward: 5′-AAG CTA GTC CGA AGG GGT TG-3′; Sepp1a reverse: 5′-TGC CCT CTG AGT TTA GCA TTG-3′. Sepp1b forward: 5′-CGT CCG TCT TCC CTC AGA C-3′; Sepp1b reverse: same as Sepp1a reverse. Sepp1c forward: 5′-GAT CGG AAA CTG TTC AGG GGC TT-3′; Sepp1c reverse: 5′-TCT CCA CAT TGC TGA GGT TGT CCT C-3′. 18SrRNA forward: 5′-CGA TTG GAT GGT TTA GTG AGG-3′; 18SrRNA reverse: 5′-AGT TCG ACC GTC TTC TCA GC-3′. RNA was harvested from N2A cells and mouse tissue using TRIzol reagent (Invitrogen), and cDNA was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to manufacturer's instructions. qPCR reactions contained cDNA, oligonucleotide primers and Platinum SYBR Green qPCR SuperMix (Invitrogen). qPCR reactions were run on a Roche LightCycler 480 II.
5′-RACE
We performed 5′-RACE using a 5′/3′ 2nd Generation RACE kit (Roche) according to the manufacturer’s instructions. RNA was extracted from mouse neonatal (postnatal day 7) brain, liver, heart, and kidney and from adult (6 mo) male or female brain, and RNA extracted with TRIzol reagent (Invitrogen) per manufacturer’s instructions. First, cDNA was synthesized from a primer binding to the start of exon 5 having the sequence 5′- CAC AGT TTT ACA GAA GTC TTC ATC TTC-3′. After adding a poly-A tail to the 3′ end of the cDNA, a PCR was performed using the kit’s 5′ oligo-dT anchor primer and a reverse primer binding to exon 2 with the sequence 5′-TGC CCT CTG AGT TTA GCA TTG-3′. The PCR product was diluted 1:20, and a subsequent PCR reaction was performed using the kit’s anchor primer and either the previous reverse primer, or primers targeted to the 3 predicted transcript variants: Sepp1a, 5′-TTG CTG AGG TTG TCG CAA CCC CTT-3′, Sepp1b, 5′-AGG TTG TCT GAG GGA AGA CGG ACG-3′, and Sepp1c, 5′-TGT CCT CGC CCT CTG AGG TGC A-3′. All PCR reactions were performed using Deep Vent (exo-) polymerase (New England BioLabs) in ThermoPol reaction buffer (NEB). The final PCR products were separated on 1% agarose gels, and DNA purified from excised bands using kit components. Products were cloned using the Topo II cloning kit (Invitrogen) and sequenced.
Cell culture
N2A neuroblastoma cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Gibco) and differentiated in Neurobasal Medium (Gibco) containing B27 supplement (Gibco) for 48 h before transfection, harvest or flow cytometry experiments.
siRNA
siRNAs targeting total Sepp1 and each transcript variant and non-targeting siRNA were purchased from Ambion. Total Sepp1 sense: 5′-GCC AUU AAG AUC GCU UAC Utt-3′, total Sepp1 antisense: 5′-AGU AAG CGA UCU UAA UGG Ctt-3′. siRNA transfections were conducted using Lipofectamine 2000 reagent (Invitrogen) according to manufacturer's instructions. Media was collected and RNA was harvested from transfected cells 24 h after transfection. Sepp1 was purified from culture media by first precipitating with 75% saturated ammonium sulfate followed by centrifugation at 5000 G for 30 min. Pellets were resuspended in binding/wash buffer (100 mM sodium phosphate pH 8, 600 mM NaCl, 0.02% Tween-20). Sepp1 was pulled down using Dynabeads His-tag Isolation and Pulldown magnetic beads (Invitrogen) per manufacturer’s instructions. Sepp1 eluted with imidazole solution (300 mM Imidazole, 50 mM Sodium phosphate pH 8, 300 mM NaCl 0.01% Tween-20) was separated by electrophoreses using 10–20% gradient Tris-HCL Criterion Precast gel (Bio-Rad Laboratories, Inc.) and transferred to Immobilon-PSQ Polyvinylidene Difluoride (PVDF) membrane (Millipore). Membranes were blocked in Odyssey blocking buffer (Li-Cor), and protein detected using a custom rabbit anti-Sepp1 antibodyCitation48 with Li-Cor infrared-fluorescent secondary antibodies detected with an Odyssey scanner. Membranes with media samples were later stained with SimplyBlue (Invitrogen) in order to normalize samples to the 67 kD albumin band.
Locked nucleic acid
Locked nucleic acid targeting miR-7 microRNA and a non-targeting control were purchased from Gene Tools (Philomath) and transfected using Arrest-in reagent (Open Biosystems) according to manufacturer's instructions. Anti-miR-7: 5′-ACA ACA AAA TCA CTA GTC TTC CA-3′; standard control: 5′-CCT CTT ACC TCA GTT ACA ATT TAT A-3′. RNA was harvested from transfected cells 24 h after transfection. Alternatively, neutralized [75Se] selenious acid (3 μCi/ml; specific activity, 1,000 Ci/mmol) was added to the culture media following transfection, followed by incubation for 20 h. Media was collected and cell lysates were prepared using CelLytic MT (Sigma Aldrich Co) according to the manufacturer’s instructions. The proteins were separated on a 10–20% gradient Tris-HCL Criterion Precast gel (Bio-Rad Laboratories, Inc.) and transferred to Immobilon-PSQ Polyvinylidene Difluoride (PVDF) membrane (Millipore), followed by phosphorimaging using a Typhoon laser scanner (GE Healthcare).
Flow cytometry
N2A cells transfected with siRNAs were stained using propidium iodine and Alexa Fluor 488 annexin V from the Vybrant Apoptosis Assay Kit #2 (Invitrogen). Samples were run on a FACSCaliber (Becton Dickinson), and fluorescence was read for 10,000 cells from each sample.
| Abbreviations: | ||
| 5′-RACE | = | rapid amplification of 5′ DNA ends |
| ApoER2 | = | apolipoprotein E receptor 2 |
| KO | = | knockout |
| LNA | = | locked nucleic acid |
| LTP | = | long-term potentiation |
| miR-10a | = | microRNA-10a |
| miR-122 | = | microRNA-122 |
| miR-7 | = | microRNA-7 |
| miRNA | = | microRNA |
| NCBI | = | National Center for Biotechnology Information |
| ORF | = | open reading frame |
| PCR | = | polymerase chain reaction |
| PVDF | = | Polyvinylidene Difluoride |
| qPCR | = | quantitative reverse transcriptase PCR |
| RNAi | = | RNA interference |
| rRNA | = | ribosomal RNA |
| Se | = | selenium |
| SECIS | = | selenocysteine insertion sequence |
| Sepp1 | = | selenoprotein P |
| siRNA | = | small interfering RNA |
| SNPs | = | single nucleotide polymorphisms |
| TRα | = | thyroid hormone receptor alpha |
| UTR | = | untranslated region |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Suguru Kurokawa for helpful suggestions and discussion, and Arjun Raman and Arlene Parubrub for manuscript review. Research supported by NIH P20 RR016467/GM103466 (FPB), NIH RO1 NS40302 (MJB), Hawaii Community Foundation Ingeborg v.F. McKee Fund 08PR-43031 (FPB) and NIH G12 RR003061/MD007601 which supports the JABSOM histology/imaging core facility. All animal procedures and experimental protocols were approved by the University of Hawaii Institutional Animal Care and Use Committee.
References
- Burk RF, Hill KE. Selenoprotein P: an extracellular protein with unique physical characteristics and a role in selenium homeostasis. Annu Rev Nutr 2005; 25:215 - 35; http://dx.doi.org/10.1146/annurev.nutr.24.012003.132120; PMID: 16011466
- Steinbrenner H, Alili L, Bilgic E, Sies H, Brenneisen P. Involvement of selenoprotein P in protection of human astrocytes from oxidative damage. Free Radic Biol Med 2006; 40:1513 - 23; http://dx.doi.org/10.1016/j.freeradbiomed.2005.12.022; PMID: 16632112
- Traulsen H, Steinbrenner H, Buchczyk DP, Klotz LO, Sies H. Selenoprotein P protects low-density lipoprotein against oxidation. Free Radic Res 2004; 38:123 - 8; http://dx.doi.org/10.1080/10715760320001634852; PMID: 15104205
- Alabi NS, Whanger PD, Wu AS. Interactive effects of organic and inorganic selenium with cadmium and mercury on spermatozoal oxygen consumption and motility in vitro. Biol Reprod 1985; 33:911 - 9; http://dx.doi.org/10.1095/biolreprod33.4.911; PMID: 4084636
- Sasakura C, Suzuki KT. Biological interaction between transition metals (Ag, Cd and Hg), selenide/sulfide and selenoprotein P. J Inorg Biochem 1998; 71:159 - 62; http://dx.doi.org/10.1016/S0162-0134(98)10048-X; PMID: 9833321
- Peters U, Chatterjee N, Hayes RB, Schoen RE, Wang Y, Chanock SJ, et al. Variation in the selenoenzyme genes and risk of advanced distal colorectal adenoma. Cancer Epidemiol Biomarkers Prev 2008; 17:1144 - 54; http://dx.doi.org/10.1158/1055-9965.EPI-07-2947; PMID: 18483336
- Bellinger FP, Raman AV, Reeves MA, Berry MJ. Regulation and function of selenoproteins in human disease. Biochem J 2009; 422:11 - 22; http://dx.doi.org/10.1042/BJ20090219; PMID: 19627257
- Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, Harney JW, et al. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature 1991; 353:273 - 6; http://dx.doi.org/10.1038/353273a0; PMID: 1832744
- Burk RF, Hill KE, Selenoprotein P. An Extracellular Protein with Unique Physical Characteristics and a Role in Selenium Homeostasis. Annu Rev Nutr 2004; PMID: 16011466
- Stoytcheva Z, Tujebajeva RM, Harney JW, Berry MJ. Efficient incorporation of multiple selenocysteines involves an inefficient decoding step serving as a potential translational checkpoint and ribosome bottleneck. Mol Cell Biol 2006; 26:9177 - 84; http://dx.doi.org/10.1128/MCB.00856-06; PMID: 17000762
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol 2008; 9:219 - 30; http://dx.doi.org/10.1038/nrm2347; PMID: 18270516
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell 2008; 132:9 - 14; http://dx.doi.org/10.1016/j.cell.2007.12.024; PMID: 18191211
- Mazière P, Enright AJ. Prediction of microRNA targets. Drug Discov Today 2007; 12:452 - 8; http://dx.doi.org/10.1016/j.drudis.2007.04.002; PMID: 17532529
- Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development 2005; 132:4645 - 52; http://dx.doi.org/10.1242/dev.02070; PMID: 16224044
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci U S A 2007; 104:9667 - 72; http://dx.doi.org/10.1073/pnas.0703820104; PMID: 17535905
- Jopling CL, Norman KL, Sarnow P. Positive and negative modulation of viral and cellular mRNAs by liver-specific microRNA miR-122. Cold Spring Harb Symp Quant Biol 2006; 71:369 - 76; http://dx.doi.org/10.1101/sqb.2006.71.022; PMID: 17381319
- Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 2008; 30:460 - 71; http://dx.doi.org/10.1016/j.molcel.2008.05.001; PMID: 18498749
- Lee I, Ajay SS, Yook JI, Kim HS, Hong SH, Kim NH, et al. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res 2009; 19:1175 - 83; http://dx.doi.org/10.1101/gr.089367.108; PMID: 19336450
- Takemoto AS, Berry MJ, Bellinger FP. Role of selenoprotein P in Alzheimer’s disease. Ethn Dis 2010; 20:Suppl 1 S1 - 92, 5; PMID: 20521393
- Lai EC, Posakony JW. The Bearded box, a novel 3′ UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development 1997; 124:4847 - 56; PMID: 9428421
- Leviten MW, Lai EC, Posakony JW. The Drosophila gene Bearded encodes a novel small protein and shares 3′ UTR sequence motifs with multiple Enhancer of split complex genes. Development 1997; 124:4039 - 51; PMID: 9374401
- Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev 2005; 19:1067 - 80; http://dx.doi.org/10.1101/gad.1291905; PMID: 15833912
- Jackson RJ, Standart N. How do microRNAs regulate gene expression?. Sci STKE 2007; 2007:re1; http://dx.doi.org/10.1126/stke.3672007re1; PMID: 17200520
- Schomburg L, Riese C, Renko K, Schweizer U. Effect of age on sexually dimorphic selenoprotein expression in mice. Biol Chem 2007; 388:1035 - 41; http://dx.doi.org/10.1515/BC.2007.128; PMID: 17937617
- Raman AV, Pitts MW, Seyedali A, Hashimoto AC, Seale LA, Bellinger FP, et al. Absence of selenoprotein P but not selenocysteine lyase results in severe neurological dysfunction. Genes Brain Behav 2012; 11:601 - 13; http://dx.doi.org/10.1111/j.1601-183X.2012.00794.x; PMID: 22487427
- Takahashi Y, Hursting SD, Perkins SN, Wang TC, Wang TT. Genistein affects androgen-responsive genes through both androgen- and estrogen-induced signaling pathways. Mol Carcinog 2006; 45:18 - 25; http://dx.doi.org/10.1002/mc.20153; PMID: 16299812
- Méplan C, Crosley LK, Nicol F, Beckett GJ, Howie AF, Hill KE, et al. Genetic polymorphisms in the human selenoprotein P gene determine the response of selenoprotein markers to selenium supplementation in a gender-specific manner (the SELGEN study). FASEB J 2007; 21:3063 - 74; http://dx.doi.org/10.1096/fj.07-8166com; PMID: 17536041
- Peters MM, Hill KE, Burk RF, Weeber EJ. Altered hippocampus synaptic function in selenoprotein P deficient mice. Mol Neurodegener 2006; 1:12; http://dx.doi.org/10.1186/1750-1326-1-12; PMID: 16984644
- Burk RF, Hill KE, Olson GE, Weeber EJ, Motley AK, Winfrey VP, et al. Deletion of apolipoprotein E receptor-2 in mice lowers brain selenium and causes severe neurological dysfunction and death when a low-selenium diet is fed. J Neurosci 2007; 27:6207 - 11; http://dx.doi.org/10.1523/JNEUROSCI.1153-07.2007; PMID: 17553992
- Olson GE, Winfrey VP, Nagdas SK, Hill KE, Burk RF. Apolipoprotein E receptor-2 (ApoER2) mediates selenium uptake from selenoprotein P by the mouse testis. J Biol Chem 2007; 282:12290 - 7; http://dx.doi.org/10.1074/jbc.M611403200; PMID: 17314095
- Rogers JT, Weeber EJ. Reelin and apoE actions on signal transduction, synaptic function and memory formation. Neuron Glia Biol 2008; 4:259 - 70; http://dx.doi.org/10.1017/S1740925X09990184; PMID: 19674510
- Weeber EJ, Beffert U, Jones C, Christian JM, Forster E, Sweatt JD, et al. Reelin and ApoE receptors cooperate to enhance hippocampal synaptic plasticity and learning. J Biol Chem 2002; 277:39944 - 52; http://dx.doi.org/10.1074/jbc.M205147200; PMID: 12167620
- Skalsky RL, Cullen BR. Reduced expression of brain-enriched microRNAs in glioblastomas permits targeted regulation of a cell death gene. PLoS One 2011; 6:e24248; http://dx.doi.org/10.1371/journal.pone.0024248; PMID: 21912681
- Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, et al. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res 2008; 68:3566 - 72; http://dx.doi.org/10.1158/0008-5472.CAN-07-6639; PMID: 18483236
- Mouradian MM. MicroRNAs in Parkinson’s disease. Neurobiol Dis 2012; 46:279 - 84; http://dx.doi.org/10.1016/j.nbd.2011.12.046; PMID: 22245218
- Chen H, Shalom-Feuerstein R, Riley J, Zhang SD, Tucci P, Agostini M, et al. miR-7 and miR-214 are specifically expressed during neuroblastoma differentiation, cortical development and embryonic stem cells differentiation, and control neurite outgrowth in vitro. Biochem Biophys Res Commun 2010; 394:921 - 7; http://dx.doi.org/10.1016/j.bbrc.2010.03.076; PMID: 20230785
- Zhang Y, Zhou Y, Schweizer U, Savaskan NE, Hua D, Kipnis J, et al. Comparative analysis of selenocysteine machinery and selenoproteome gene expression in mouse brain identifies neurons as key functional sites of selenium in mammals. J Biol Chem 2008; 283:2427 - 38; http://dx.doi.org/10.1074/jbc.M707951200; PMID: 18032379
- Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, et al. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci 2006; 9:99 - 107; http://dx.doi.org/10.1038/nn1618; PMID: 16369481
- Burk RF, Hill KE. Selenoprotein P-expression, functions, and roles in mammals. Biochim Biophys Acta 2009; 1790:1441 - 7; http://dx.doi.org/10.1016/j.bbagen.2009.03.026; PMID: 19345254
- Chiu-Ugalde J, Theilig F, Behrends T, Drebes J, Sieland C, Subbarayal P, et al. Mutation of megalin leads to urinary loss of selenoprotein P and selenium deficiency in serum, liver, kidneys and brain. Biochem J 2010; 431:103 - 11; http://dx.doi.org/10.1042/BJ20100779; PMID: 20653565
- Olson GE, Winfrey VP, Hill KE, Burk RF. Megalin mediates selenoprotein P uptake by kidney proximal tubule epithelial cells. J Biol Chem 2008; 283:6854 - 60; http://dx.doi.org/10.1074/jbc.M709945200; PMID: 18174160
- Bockmühl Y, Murgatroyd CA, Kuczynska A, Adcock IM, Almeida OF, Spengler D. Differential regulation and function of 5′-untranslated GR-exon 1 transcripts. Mol Endocrinol 2011; 25:1100 - 10; http://dx.doi.org/10.1210/me.2010-0436; PMID: 21527501
- Kreth S, Ledderose C, Kaufmann I, Groeger G, Thiel M. Differential expression of 5′-UTR splice variants of the adenosine A2A receptor gene in human granulocytes: identification, characterization, and functional impact on activation. FASEB J 2008; 22:3276 - 86; http://dx.doi.org/10.1096/fj.07-101097; PMID: 18541693
- Okada M, Nakajima K, Yaoita Y. Translational regulation by the 5′-UTR of thyroid hormone receptor α mRNA. J Biochem 2012; 151:519 - 31; http://dx.doi.org/10.1093/jb/mvs026; PMID: 22406405
- Pantopoulos K. Iron metabolism and the IRE/IRP regulatory system: an update. Ann N Y Acad Sci 2004; 1012:1 - 13; http://dx.doi.org/10.1196/annals.1306.001; PMID: 15105251
- Chen CH, Lin HY, Pan CL, Chen FC. The genomic features that affect the lengths of 5′ untranslated regions in multicellular eukaryotes. BMC Bioinformatics 2011; 12:Suppl 9 S3; http://dx.doi.org/10.1186/1471-2105-12-S9-S3; PMID: 22152105
- Kosik KS, Krichevsky AM. The Elegance of the MicroRNAs: A Neuronal Perspective. Neuron 2005; 47:779 - 82; http://dx.doi.org/10.1016/j.neuron.2005.08.019; PMID: 16157272
- Seale LA, Hashimoto AC, Kurokawa S, Gilman CL, Seyedali A, Bellinger FP, et al. Disruption of the selenocysteine lyase-mediated selenium recycling pathway leads to metabolic syndrome in mice. Mol Cell Biol 2012; 32:4141 - 54; http://dx.doi.org/10.1128/MCB.00293-12; PMID: 22890841