Abstract
We report strong somatic and germ line expression of LINE RNAs in eight different tissues of rat by using a novel ~2.8 kb genomic PstI-LINE DNA (P1-LINE) isolated from the rat brain. P1-LINE is present in a 93 kb LINE-SINE-cluster in sub-telomeric region of chromosome 12 (12p12) and as multiple truncated copies interspersed in all rat chromosomes. P1-LINEs occur as inverted repeats at multiple genomic loci in tissue-specific and mosaic patterns. P1-LINE RNAs are strongly expressed in brain, liver, lungs, heart, kidney, testes, spleen and thymus into large to small heterogeneous RNAs (~5.0 to 0.2 kb) in tissue-specific and dynamic patterns in individual rats. P1-LINE DNA is strongly methylated at CpG-dinucleotides in most genomic copies in all the tissues and weakly hypomethylated in few copies in some tissues. Small (700–75 nt) P1-LINE RNAs expressed in all tissues may be possible precursors for small regulatory RNAs (PIWI-interacting/piRNAs) bioinformatically derived from P1-LINE. The strong and dynamic expression of LINE RNAs from multiple chromosomal loci and the putative piRNAs in somatic tissues of rat under normal physiological conditions may define functional chromosomal domains marked by LINE RNAs as long noncoding RNAs (lncRNAs) unrestricted by DNA methylation. The tissue-specific, dynamic RNA expression and mosaic genomic distribution of LINEs representing a steady-state genomic flux of retrotransposon RNAs suggest for biological role of LINE RNAs as long ncRNAs and small piRNAs in mammalian tissues independent of their cellular fate for translation, reverse-transcription and retrotransposition. This may provide evolutionary advantages to LINEs and mammalian genomes.
Introduction
Mammalian genomes, for example the human genome, mouse genome and rat genome contain about 40–50% repetitive DNA sequences and 37–45% mobile genetic elements (transposons) including 17–23% long interspersed nuclear elements (LINEs).Citation1-Citation3 LINEs and SINEs (short interspersed nuclear elements) constitute the retrotransposons of mammalian genomes, which play a major role in genome evolution through their RNA-intermediates by a “copy and paste” mechanism.Citation4,Citation5 LINEs (L1s) are non-long-terminal repeat (LTR), autonomous, retrotransposons expressed as polyadenylated LINE-RNAs by RNA polymerase II and cellular transcription factors.Citation6 The full-length L1 (6–7 kb) consists of a 5′-untranslated region (5′UTR) with an internal RNA polymerase II-promoter, open reading frame (ORF)-1, ORF2, 3′UTR and a poly(A)-tail.Citation7 The ORF-1 encodes a RNA-binding protein (ORF1p), while ORF2 encodes a protein (ORF2p), which has endonuclease (EN) and reverse transcriptase (RT) activities.Citation8 The full-length L1-members with active ORF1 and ORF2 are retrotransposition-competent, however, more than 90% of L1s are retrotransposition-defective due to 5′-truncation and mutations in ORF2.Citation9 Most of the L1 sequences present in mammalian genomes are 5′-truncated because the ORF2 protein involved in the reverse transcription of L1 RNA is less efficient and is unable to copy the 5′-end of the RNA.Citation10
L1 sequences are major retrotransposons in mammalian genomes, with variability in abundance, copy number and sequences at different genomic loci.Citation11 L1s are prevalent in AT-rich, gene-poor (intergenic) regions, low-recombination regions and introns, usually in antisense orientation with respect to the nearest gene as well as in clusters, e.g., in the human X chromosome.Citation12,Citation13 Distribution of L1s in the genome differs with respect to their size and evolutionary age.Citation11 Full-length L1s are generally present in abundance in sex chromosomes whereas truncated elements are more abundant in autosomes. On evolutionary scale, young L1s are present closer to the genes, while older elements are present in gene-deficient regions or farther from the genes. About 6,00,000 copies of L1s comprise 23% of the rat (Rattus norvegicus) genome representing the autonomous, non-LTR retrotransposons.Citation3 The basic structure of the reported full-length rat L1 (RnL1, accession no DQ100473) is similar to that of mammalian L1 in structure except for the 5′UTR, which is bipartite and contains a specific tether sequence (21 bp sequence connecting the 5′UTR to ORF-1).Citation14 At the sequence level, the 3′UTR defines six rat-specific L1-subfamilies, which cover 12% of the genome and is represented by 1,50,000 copies.
About only 1.2% of mammalian genome in the form of exons is used to code for amino acids in proteins while the rest > 98% is non-protein coding in nature and transcribed into noncoding RNAs (ncRNAs). Recently, existence and functional significance of ncRNAs from mammalian genome have been reviewed.Citation15 Long ncRNAs have been described with many overlapping functions. Mammalian ncRNAs function as regulators of gene expression and assembly of cellular structures and the possible mechanisms involved in these have been described.Citation16 For examples, the lncRNA, MALAT1 regulates alternative splicing of mRNAs by regulating the SR (splice regulator) proteins. Chromatin-mediated local gene silencing by cis-acting lncRNAs, e.g., Xist RNA, which is associated with the inactivation of mammalian X chromosome; trans-(long intergenic ncRNA) lincRNA, e.g., p53-regulated lincRNA-p21 hitting multiple targets; HOTAIR lncRNA acting as scaffolds for histone modifiers; the neuronal enhancer associated lncRNAs (encRNAs) organizing enhancer activity and higher order chromatin structures; genomic reprogramming of induced pluripotent stem cells (iPSCs) by lncRNAs and nucleation of nuclear structures by lncRNAs are some other examples of functions of mammalian long ncRNAs.Citation17-Citation19 Similarly, many lncRNAs have been described as signals for cellular regulations, e.g., lincRNA-p21 and PANDA induced by DNA damage; induction of COLDAIR and COOLAIR lncRNAs by cold; ROR lincRNA colocalized with the pluripotency transcription factors like Oct4, Sox2 and Nanog. The repeat detector ncRNAs: e.g., RNA decay by Staufen; ncRNA decoys, e.g., DHFR minor, telomere associated TERRA, PANDA binding with NF-YA and Gas5-suppressing glucocorticoid receptor, MALAT1 and miRNA target mimics for miRNAs and splicing factors; ncRNA as guides, e.g., lncRNAs binding to protein factors like polycomb, MLL, TFIIB and ncRNA as scaffolds, e.g., HOTAIR, ANRIL, α-satellite repeat-binding lncRNA have been reported. These ncRNAs have been variously described to explain the mechanisms of their actions and function(s).Citation20 Biological functions of ncRNAs in the mammalian central nervous system (CNS) has been highlighted with respect to miRNAs, small nucleolar RNAs, retrotransposons, NRSE small modulatory RNAs like BC1/BC200 correlating the complexity of CNS with biogenesis, dynamics of action and combinatorial regulations of ncRNAs.Citation21 Similarly, role of ncRNAs has been explained in the neuronal network with special reference to REST/NRSF and neurological disorders.Citation22 Role of lncRNAs has also been highlighted in cancer and neurological diseases.Citation23,Citation24
Role of lncRNAs has also been explained in epigenetic regulation of chromatin and gene expression during embryological and neural differentiation, where lncRNA can function to make sequence specific recognition with promoter/enhancer of a given gene and simultaneously act as an adaptor to bind and deliver chromatin modifying protein(s) to the nucleosome(s).Citation25 Further, RNA signaling has been proposed to play a central cellular signaling role in multicellular ontogeny, homeostasis and epigenetic memory.Citation25 Long ncRNAs have also been described as regulators and integrators of gene regulatory transcriptional and RNA processing networks in multicellular organisms.Citation26
RNA sequencing and ENCODE analyses have shown an enormously large number (hundreds of thousands) of intragenic and intergenic lncRNAs catalogoued as annotated lncRNAs, which have been conserved through evolution for controlling various functions in diverse organisms such as epigenetic regulation of gene expression and linked to diseases like cancer and neurodegeneration.Citation27 It is estimated by full-length cDNA sequencing and genome-wide hybridizations that about 63% of the mouse genome sequence is pervasively transcribed, thus thousands of novel protein-coding transcripts and > 30,000 lncRNAs of intronic, intergenic and antisense nature have been described.Citation28-Citation30 Similarly, by whole chromosome RNA analysis it has been observed that about 93% of the human genome sequence is pervasively transcribed in one or the other cell type.Citation31-Citation35 Maximum of these transcripts are logically expected to be the “dark matter” of the genome or ncRNAs of diverse biological functions.Citation36
Recently, a class of small regulatory RNAs called piRNAs (PIWI-interacting RNAs) have been reported to function for silencing of transposons in the germline.Citation37-Citation40 PIWIs are germ-line specific Argonaute class of RNA-interference effector proteins associated with piRNAs. Loss of MIWI2, a mouse PIWI-homolog, leads to increased germ-line transposon activity and germ-line stem cell and meiotic defects.Citation41 Genomic DNA regions coding for piRNAs exist as large clusters (~20–100 kb) in mammalian genome and they are transcribed into precursor piRNAs (~10 kb), which are further processed into piRNAs (24–31 nt). Genomic antisense piRNA has complementary sequence with LINE-1 transcript.Citation42 RNA processing and site-specific cleavage of complementary base pairing of antisense piRNA and LINE-1 RNA complex facilitates generation of sense piRNA from the LINE-1 RNA, which again can base pair with the antisense piRNA to amplify piRNAs in a cycle of events.Citation43 Recently, expression of mammalian piRNAs outside of germ-line tissue has also been demonstrated.Citation44
We report a novel ~2.8 kb genomic LINE1, named P1-LINE (accession number GQ244317) mostly composed of ORF2 sequence and present in all chromosomes in mosaic patterns in the rat tissues. It shows high levels of heterogeneous, tissue-specific and dynamic RNA expression in individual rats. P1-LINE is mostly hypermethylated in all eight tissues studied, thus showing high levels of LINE RNA expression unrestricted by DNA methylation. P1-LINE shows five piRNAs and their possible precursor RNAs. This represents global LINE RNA expression as noncoding RNAs in somatic tissues.
Results
Cloning of LINE DNA from rat genome and sequence analysis
Since rat genome contains very high amounts of repetitive DNA (41%) and LINE sequences (23%), genomic DNA from the rat brain was digested by BamHI, EcoRI, Hind III, PstI and the DNA fragments were resolved by agarose gel electrophoresis and the six DNA fragments: 5.6 kb BamHI (B1), 2.3 kb EcoRI (E1), 1.3 kb EcoRI (E2), 3.5 kb Hind III (H1), 2.8 kb PstI (P1) and 2.3 kb PstI (P2) were gel-purified (). As an example, digestion of the rat brain genomic DNA by PstI generating the 2.8 kb and 2.3 kb DNA fragments is shown (). The 2.8 kb PstI fragment was cloned into pBluescript vector at PstI site to generate pP1 clone (). The other five DNA fragments were similarly cloned into the respective restriction enzyme sites in the pBluescript vector generating the pB1, pE1, pE2, pH1 and pP2 clones (; Fig. S1A–F).
Table 1. Genomic DNA fragments cloned from the rat brain
Figure 1. P1-LINE DNA from the rat genome. (A) Rat brain genomic DNA was completely digested with PstI (P) and resolved in 1% agarose-TAE gel. The 2.8 kb DNA band was gel-purified and cloned into PstI site of pBlueScript II SK (+) vector. (B) The clone was digested by PstI to verify the insert. V, vector [pBS-SK(+)II]; P1, insert (P1-LINE). (C) Schematic diagram of cloned P1-LINE showing LINE1, LTR/MaLR, simple repeat sequences and putative open reading frames (ORFs) of 309 aa and 212 aa, homologous to the ORF2 of RnL1. The three CCGG sites are mentioned. The five rat-specific piRNAs are piR1 (1706–1734 nt), piR2 (2364–2338 nt), piR3 (2442–2471 nt), piR4 (2451–2479 nt) and piR5 (2513–2483 nt). The 310 (2421–2730) bp probe was used for the alkaline-agarose Southern blot (Fig. 6) and denaturing northern blot for small RNA (Fig. 7). MaLR = mammalian apparent LTR (long-terminal repeat)-retrotransposon.
![Figure 1. P1-LINE DNA from the rat genome. (A) Rat brain genomic DNA was completely digested with PstI (P) and resolved in 1% agarose-TAE gel. The 2.8 kb DNA band was gel-purified and cloned into PstI site of pBlueScript II SK (+) vector. (B) The clone was digested by PstI to verify the insert. V, vector [pBS-SK(+)II]; P1, insert (P1-LINE). (C) Schematic diagram of cloned P1-LINE showing LINE1, LTR/MaLR, simple repeat sequences and putative open reading frames (ORFs) of 309 aa and 212 aa, homologous to the ORF2 of RnL1. The three CCGG sites are mentioned. The five rat-specific piRNAs are piR1 (1706–1734 nt), piR2 (2364–2338 nt), piR3 (2442–2471 nt), piR4 (2451–2479 nt) and piR5 (2513–2483 nt). The 310 (2421–2730) bp probe was used for the alkaline-agarose Southern blot (Fig. 6) and denaturing northern blot for small RNA (Fig. 7). MaLR = mammalian apparent LTR (long-terminal repeat)-retrotransposon.](/cms/asset/6958e7aa-8bcd-42f9-af52-5cc26e61aefc/krnb_a_10922402_f0001.gif)
The six DNA fragments were sequenced and the sequences were analyzed by bioinformatics tools available online and through the genome database. The four LINE DNA fragments: B1, E1, H1 and P1 were identified on the basis of percentage of repeats and LINE sequences (Fig. S1G; ; ). B1 contains 28.11% repeats and 7.08% LINE with a long-terminal repeat (LTR) at 1151–1319 bp, E1 contains 80.09% LINE as the sole repeat sequence, H1 contains 65.40% repeats and 43.34% LINE, while P1 contains 98.03% repeats and 92.52% LINE. E2 contains 1.55% GC-rich repeat and P2 contains 3.47% simple repeats, 5.74% long-terminal repeat (LTR) and both are derived from the intergenic regions. Since, P1-LINE DNA contains 98% repeats and 92% LINE, it was studied further. It is a 2796 bp repetitive DNA containing 92% LINE-1 (L1) sequences with a LTR (23–108 bp) and simple repeats (1163–1230 bp). P1-LINE showed homology with ORF2 of the LINE isolated from Rattus norvegicus (RnL1, accession number DQ100473) in the following regions: 123–1162 bp, 1231–1404 bp and 1424–2796 bp of P1-LINE with 4328–5408 bp, 4164–4332 bp and 4962–5859 bp of RnL1, respectively ().Citation14 The ORF2 homologous sequences of P1-LINE (123–1013 bp and 2157–2792 bp) can encode for a 309 aa and a 212 aa partial ORF2 proteins in the -2 frame, respectively. The rest of the ORF2 sequence is mutated, hence it may not code for the protein. Part of ORF2 is duplicated (123–562 bp and 2347–2794 bp) in P1-LINE. The 2.8 kb P1-LINE is located in the 12p12 locus of rat chromosome 12, which is a sub-telomeric region ().
Figure 2. Presence of P1-LINE sequences in the rat genome. (A) The 93 kb region (accession number NW_047365.1) of the rat chromosome 12 (12p12, 1–93197 bp) shows a cluster of 53 LINEs (1–53) as light gray boxes in the middle bar and 57 SINEs (1–57) as dark gray boxes in two parts (1–22 and 23–57) in the top (1–46345 bp) and bottom (46346–93197 bp) bars, respectively. The full length L1 (4 and 13) sequences are marked with a star. P1-LINE is drawn as a black solid line from 35 to 37 in the middle bar. (B) Distribution and abundance of P1-LINE sequences in the rat chromosomes ().
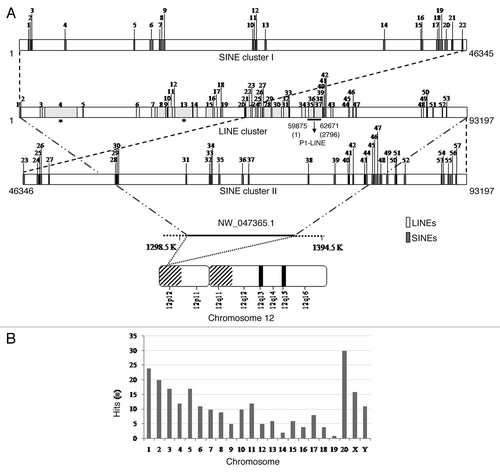
BLAT analysis shows that the 1400–2796 bp region of P1-LINE is present in all rat chromosomes, at a number of loci, in more than hundred thousand copies (; ). We named this region as P1-LINE common region (P1-LINEc). Only selected homology-hits with a minimum score of 1000 and identity of 90% are shown in . BLAT analysis also showed more number of hits on the bigger rat chromosomes, like the chromosomes 1, 2, 3, 4, 5 and X, while it decreased as the size of the chromosomes decreases, except for the chromosomes 10, 11 and 20. These observations indicate that P1-LINE is present throughout the rat genome in multiple copies. We analyzed a 93 kb region of 12p12 locus of chromosome 12 (accession number NW_047365.1) having P1-LINE by repeat masker (). Cluster analysis showed presence of 53 LINEs (1–53), out of which, two are full length LINEs of 6402 bp and 6166 bp (no. 4 and 13) marked with asterisks and the rest are truncated copies. P1-LINE is shown as a black bar at position 35–37. In the same 93 kb region, 57 SINEs are also present. The length of the SINEs varies from 12 to 303 bp. Thus, it is a very rich zone of LINE-SINE cluster in the sub-telomeric region of chromosome 12. The Promoter 2.0 prediction showed no promoter sequence in P1-LINE but several putative transcription factor binding sites were detected in the 2.8 kb sequence by using PROMO 3.0 program within a dissimilarity margin of less than or equal to 5%. Some of these transcription factors are C/EBPα, C/EBPβ, c-Fos, YY1, GR, Pax-4a.
Table 2. P1-LINE DNA in rat chromosomes
P1-LINE sequence was analyzed with the piRNA database to look for presence of putative piRNA(s). Interestingly, P1-LINE showed homology with five rat piRNAs of 27–31 nt in the 1706–2513 bp ORF2 region (). Out of the five putative piRNAs, three are generated from the top strand, i.e., piR1 (1706–1734 nt), piR3 (2442–2471 nt) and piR4 (2451–2479 nt), while two piRNAs are generated from the bottom strand, i.e., piR2 (2364–2338 nt) and piR5 (2513–2483 nt), respectively. The five piRNAs are 100% identical to P1-LINE. The piR1 to piR5 piRNAs exist in several thousands of copies, such as 8505, 23623, 24235, 23164 and 28700 copies in the rat genome, respectively and they have been reported from the rat testis.Citation45
Table 3. Putative piRNAs from P1-LINE in rat genome
P1-LINE RNA expression in rat tissues by northern blot analysis
We investigated P1-LINE RNA expression in eight different tissues of the adult rat under normal physiological conditions by northern blot analysis using 32P-labeled 2.8 kb P1-LINE DNA probe. It shows strong P1-LINE RNA expression in multiple tissues of the rat, such as brain, liver, lungs, heart, kidney, testis, spleen and thymus (; Fig. S2A and B). The LINE RNAs are heterogeneous (> 5.0 to 0.2 kb) in size, their expression levels are different in different tissues and their expression pattern is variable in four individual rats (; Fig. S2A and B), which was quantitated by densitometry and represented as relative RNA expression, i.e., P1-LINE RNA normalized to 28S rRNA (). It shows highest expression in the heart and testes; high expression in the lungs, kidney, spleen and thymus followed by brain and liver. Based on the densitometry of the northern blot signals, shows the variations in the RNA expression in the tissues of four individual rats in a scale of weak (< 3), moderate (3–4), strong (4–5), very strong (> 5). The LINE RNA expression is very strong in the heart, kidney, testis, spleen and thymus; strong in the brain and lungs and moderate in the liver. With respect to individual variations in the expression level, testis is highly variable; brain, liver, lungs, heart and kidney show low variability; while spleen and thymus show more or less no variation. However, all the tissues show variations with respect to the size of the transcripts. Lungs, spleen and thymus show strong RNA expression ranging from large to small size transcripts. In case of lungs, two out of four rats show large transcripts, whereas all four show small transcripts. Spleen and thymus show mainly six major types of transcripts in addition to the heterogeneous transcripts with more or less same pattern of expression in all four individual rats. The heterogeneous LINE RNAs () must be produced from the 5′truncated P1-LINEs present in multiple copies at different genomic sites (), hence they represent long non-protein coding RNAs (lncRNAs). We also examined RNA expression from the other 5 repetitive DNAs. The E1 repetitive DNA (80% LINE) shows strong RNA expression, H1 (43% LINE) shows moderate RNA expression, B1 (7% LINE) and P2 (no LINE) show weak RNA expression in some tissues (Fig. S2C–F) but E2 (no LINE) shows no RNA expression (data not shown). Thus there is a correlation between the level of RNA expression and the content of LINE sequence of the genomic repetitive DNAs. The strong and heterogeneous P1-LINE RNA expression in the rat tissues represents LINEs expressing from multiple chromosomal sites in the rat genome. We also checked P1-LINE RNA expression in the eight tissues of 4 week-old immature rat, which also showed expression of LINE RNAs similar to that of the adult rat tissues (Fig. S3A). Thus, P1-LINE showed heterogeneous, tissue-specific and dynamic patterns of endogenous LINE RNA expression in seven somatic tissues and the male gonadal tissue (testis) of the immature and adult rats under normal physiological conditions. Therefore P1-LINE sequences in the rat genome show widespread expression of noncoding RNAs (ncRNAs) in the somatic and gonadal (testes) tissues of the rat.
Figure 3. Expression of P1-LINE RNA in adult rat tissues by northern blot. RNA expression of P1-LINE in eight different tissues of adult rat was examined by northern blot hybridization using 32P-labeled 2.8 kb P1-LINE DNA probe. Heterogeneous RNAs of ~5.0 to 0.2 kb sizes were detected. Some distinct transcripts are marked with arrows. 100 pg of the unlabelled probe was used as a positive control (+) for hybridization. Equal loading of total RNA in the gel is shown by methylene blue staining of 28S and 18S rRNAs. Expression of 28S rRNA is shown as an internal reference. (A) and (B) represent two experiments showing RNA expression for two out of four individual rats, Figure S2A and Brepresent the other two rats. Densitometric quantitation of P1-LINE RNA normalized to 28S rRNA is represented as relative RNA expression. (C) Relative RNA expression of P1-LINE (Mean ± SEM, see Table 4) from the four rats.
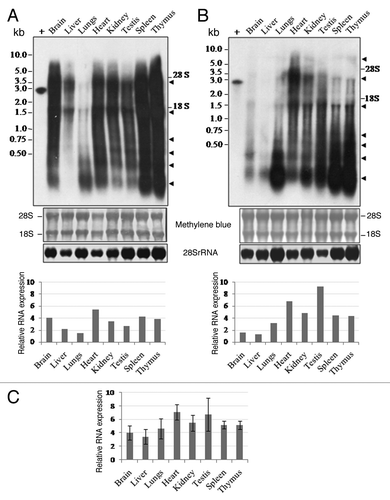
Table 4. Level of P1-LINE RNA expression and size of the transcripts
P1-LINE RNA expression in rat tissues by RT-PCR analysis
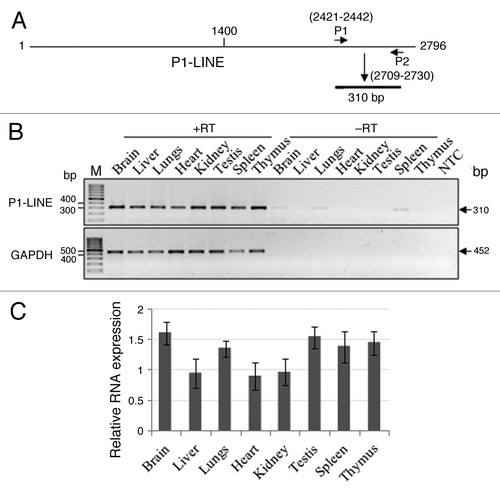
Since almost all P1-LINE copies distributed throughout the rat genome contain the P1-LINE common (P1-LINEc) region, primers (P1 and P2) were designed from this region to study its RNA expression and genomic organization (). P1-LINE RNA expression measured by RT-PCR was normalized to expression of GAPDH mRNA. It shows that the brain (1.602 ± 0.181), testes (1.538 ± 0.176), thymus (1.447 ± 0.190), spleen (1.385 ± 0.255) and lungs (1.345 ± 0.130) have higher expression levels whereas the kidney (0.966 ± 0.216), liver (0.950 ± 0.234) and heart (0.901 ± 0.220) show relatively lower expression levels (). This represents total P1-LINE RNA expression from the rat genome in the tissues. The results from the northern blot hybridization and RT-PCR both show that P1-LINE RNAs are expressed in the tissues. However, the results obtained by these two techniques are not similar, which may be due to differences in the presence of the 2.8 kb P1-LINE and the 310 bp P1-LINEc regions in the genome due to variations in the truncated P1-LINE copies, their copy no. as well as their transcriptional activity in the tissues. The truncated copies of P1-LINE are different in different tissues and rats with respect to the extent of truncation. We expect their transcriptional activity by RNA polymerase II may be different. The RT-PCR would amplify from the RNAs from all the truncated P1-LINE copies as well as the 2.8 kb P1-LINE. However for the northern blot, the extent of hybridization of the RNAs from the truncated P1-LINE copies and the 2.8 kb P1-LINE may be different (the probe being the 2.8 kb P1-LINE DNA). This may contribute to variations in the results from RT-PCR and northern blot experiments.
Figure 4. Expression of P1-LINE RNA in adult rat tissues by RT-PCR. RNA expression of P1-LINE in eight different tissues of adult rat was examined by RT-PCR. (A) P1 and P2 primers from the P1-LINE common region homologous to ORF2 generate a 310 bp amplicon. (B) Representative expression of P1-LINE RNA in eight different tissues. RT, reverse transcriptase; -RT, no RT negative control; NTC, no template (1st strand cDNA) negative control; GAPDH (452 bp), reference signal. (C) Relative RNA expression (P1-LINE RNA /GAPDH mRNA) by densitometric quantitation of the results from four individual rats.
P1-LINE is highly methylated in rat genome
LINE sequences are reported to be methylated in differentiated cells to silence their expression under normal conditions. Since we observed high levels of RNA expression for P1-LINE in somatic tissues of the rat under normal conditions, this prompted us to check the methylation status of P1-LINE in the genome. We analyzed the DNA methylation status of P1-LINE by PstI+Msp I/HpaII restriction digestions of the genomic DNA from the tissues followed by Southern blot hybridization by the 32P-labeled 2.8 kb P1-LINE DNA probe (). We observed that HpaII-digested P1-LINE DNA produced large heterogeneous fragments of > 10 kb to 2 kb, but in addition to this, Msp I digested the DNA to produce six major fragments of 5.0, 3.0, 2.8, 1.3, 0.89 and 0.4 kb. The 2.8, 1.3, 0.89 and 0.4 kb fragments were strongly produced by Msp I, but not by HpaII, in all the eight tissues (brain, liver, lungs, heart, kidney, testes, spleen and thymus), hence they were methylated at the internal C of 5′CCGG3′ sequence present at the three positions (1117, 1485 and 1903) in the 2.8 kb P1-LINE (). The 0.89 and 0.4 kb fragments are also produced from the cloned 2.8 kb P1-LINE DNA (). A 5.0 kb MspI fragment is strongly produced in the lungs, thymus and weakly produced in other tissues. The weak HpaII signals at 3.0 kb, 1.3 kb and 0.89 kb in the heart and at 3.0 kb, 2.8 kb, 1.3 kb, 0.89 kb and 0.4 kb in the testis show hypomethylated P1-LINE sequences. This suggests tissue-specific P1-LINE methylation in rat genome. Quantitation of the four Msp I DNA fragments (2.8 kb, 1.3 kb, 0.89 kb and 0.4 kb) by densitometry shows that in all the tissues the 2.8 kb DNA was methylated up to 50–70% whereas the 1.3, 0.89 and 0.4 kb DNAs were methylated up to about 90% (). This shows that P1-LINE is mostly hypermethylated but some hypomethylated P1-LINE sequences are also present in rat genome. The DNA methylation status of P1-LINE represents LINEs from the entire rat genome and to some extent it also shows a tissue-specific methylation pattern of LINEs. Thus P1-LINE is transcribed from the genome in a methylation-unrestricted manner.
Figure 5. Methylation of P1-LINE DNA in rat genome. (A) The cloned 2.8 kb P1-LINE DNA fragment (P1) was digested by Msp I (M)/HpaII (H) and used as a positive control to assess the corresponding genomic P1-LINE DNA fragments. (B) Genomic DNA purified from eight different tissues of adult rat was digested by PstI+Msp I (M) and PstI+HpaII (H) followed by Southern blot hybridization with 32P-labeled 2.8 kb P1-LINE DNA probe to assess DNA-methylation at 5′CCGG3′ sequences of P1-LINE in the rat genome. Four Msp I bands (2.8 kb, 1.3 kb, 0.89 kb and 0.4 kb) are marked by arrows.The blot is representative of two independent experiments from individual rats. + is purified 2.8 kb P1-LINE DNA used as positive control. (C) Densitometric quantitation of the four Msp I bands (2.8 kb, 1.3 kb, 0.89 kb and 0.4 kb) showing % methylation of P1-LINE expressed as [(Msp I-HpaII)/Msp I]x100 in the tissues.
![Figure 5. Methylation of P1-LINE DNA in rat genome. (A) The cloned 2.8 kb P1-LINE DNA fragment (P1) was digested by Msp I (M)/HpaII (H) and used as a positive control to assess the corresponding genomic P1-LINE DNA fragments. (B) Genomic DNA purified from eight different tissues of adult rat was digested by PstI+Msp I (M) and PstI+HpaII (H) followed by Southern blot hybridization with 32P-labeled 2.8 kb P1-LINE DNA probe to assess DNA-methylation at 5′CCGG3′ sequences of P1-LINE in the rat genome. Four Msp I bands (2.8 kb, 1.3 kb, 0.89 kb and 0.4 kb) are marked by arrows.The blot is representative of two independent experiments from individual rats. + is purified 2.8 kb P1-LINE DNA used as positive control. (C) Densitometric quantitation of the four Msp I bands (2.8 kb, 1.3 kb, 0.89 kb and 0.4 kb) showing % methylation of P1-LINE expressed as [(Msp I-HpaII)/Msp I]x100 in the tissues.](/cms/asset/57cc4605-a85e-4f46-853b-157e927d3766/krnb_a_10922402_f0005.gif)
IRS-PCR shows tissue-specific and mosaic patterns of P1-LINEs in rat genome
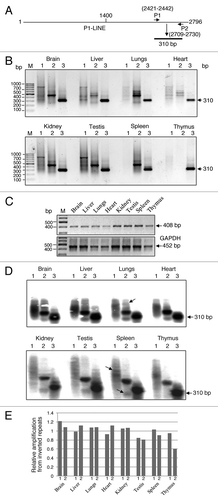
Bioinformatically, we found that P1-LINE is present in multiple copies at a number of loci and in both orientations in the rat genome. This suggested that some copies of P1-LINE should be present as inverted repeats. shows a schematic representation of PCR-amplification of the 310 bp DNA from the P1-LINEc region by P1+P2 primers. IRS-PCR () by single primer (P1 or P2) amplification shows discrete bands in almost all tissues indicating the presence of P1-LINE sequences as inverted repeats at multiple sites in rat genome. Spleen and thymus show very weak amplification by P1 primer and by P1 or P2 primer respectively. Distinct bands are observed for P1 primer in brain, liver, heart, kidney and testis, while weak bands are observed in lungs. Similarly, distinct bands are observed for P2 primer in brain, lungs, kidney, testis and spleen, while weak bands are observed in liver and heart. For P1+P2 primers, all the tissues show a strong band of 310 bp indicating amplification from all P1-LINE genomic copies. Genomic DNA-PCR of a rat repetitive DNA sequence (accession number GQ463151, upper panel) and GAPDH gene (lower panel) are used as reference signals for comparison (). The IRS-PCR products were also analyzed by alkaline-agarose gel and Southern hybridization with 32P-labeled 310 bp P1-LINEc probe (). Distinct P1-LINE specific DNA bands were observed as inverted copies amplified by either P1 or P2 primer and as total copies by P1+P2 primers. Total amplification of genomic P1-LINE sequences by P1+P2 primers detected as hybridization signals varies between 17–18% (3.1 to 2.6x105 IDV, data not shown) whereas the relative amplification signals from P1-LINE inverted repeats vary between 1.2 to 0.8 for P1 and 1.1 to 0.6 for P2 in the tissues (). This shows that the positions of the inverted copies of P1-LINE in the genome and their copy number in different tissues of the rat are different. In spleen and thymus, the inverted repeats of P1-LINE may be less in copies. Therefore, P1-LINE shows tissue-specific and mosaic distribution patterns in rat genome.
Figure 6. Interspersed-repetitive sequence-PCR (IRS-PCR) of P1-LINE in rat genome. (A) A schematic diagram showing the primers (P1 and P2) from the common region (1400–2796 bp) of P1-LINE and its expected amplification-product of 310 bp. (B) IRS-PCR products from genomic DNA from eight different rat tissues using single primer (P1 or P2) marked as lane 1 and lane 2, and double primers (P1+P2) marked as lane 3 are shown, respectively. (C) Genomic DNA-PCR of a rat repetitive DNA sequence (accession number GQ463151, upper panel) is shown as the 408 bp and the GAPDH gene (lower panel) as the 452 bp amplicons as reference signals. M- 100 bp DNA ladder used as a molecular size marker. (D) Alkaline-agarose gel electrophoresis and Southern blot hybridization of the IRS-PCR products from the tissues with the 32P-labeled 310 bp P1-LINE DNA probe. Arrow indicates a band which is absent in some other tissue. (E) Densitometric quantitation of the IRS-PCR-Southern blot signals from (D) expressed as relative amplification (lane 1/3 and 2/3) from the inverted repeats of P1-LINE.
Expression of small RNA from P1-LINE
DNA sequence analysis and homology search showed that five rat piRNAs of 27–31 nt can be generated from 1706–2513 bp (ORF2) region of P1-LINE (, ). This includes three piRNAs from the top strand (piR1, piR3 and piR4) and two piRNAs from the bottom strand (piR2 and piR5) showing their opposite orientation. The 5 putative piRNAs from P1-LINE are as follows: piR1 (1706–1734) is 29 nt and exists in 8505 copies, piR2 (2364–2338) is 27 nt and exists in 23623 copies, piR3 (2442–2471) is 30 nt and exists in 24235 copies, piR4 (2451–2479) is 29 nt and exists in 23164 copies and piR5 (2513–2483) is 31 nt and exists in 28700 copies in rat genome. All 5 piRNAs are 100% identical to the piRNAs in the database reported from the rat testes. In order to detect small RNAs (piRNAs) from P1-LINE in the tissues, denaturing polyacrylamide+urea northern blot hybridization was performed using 32P-labeled 310 bp PCR-amplified P1-LINEc DNA probe, which is derived from the common (ORF2) region. Heterogeneous transcripts of ~700 to ~75 nt were detected in all the eight tissues along with two distinct RNAs of ~400 and ~170 nt detected in some tissues (). These transcripts are also detected in the tissues of immature (4 week) rats (Fig. S3B). From the sequence analysis, we detected ESTs in the rat EST-database matching with P1-LINE up to 88–96% identity mostly representing the 5′ untranslated (5′-UTR) and 3′-UTR regions of the transcripts/mRNAs. These are 474–699 nt sequences from the transcripts/mRNAs detected in the brain, liver, spleen and 12 d fetus of the rat (). Interestingly, all these ESTs show homology with the common region (1424–2796 bp) of P1-LINE present in all rat chromosomes indicating existence of homologous sequences between the transcripts/mRNAs and the ORF2 sequence of the truncated LINEs. Therefore, the small transcripts detected by the PAGE-urea northern blot in different rat tissues may be partly homologous to these ESTs from the rat EST-database. We suggest that these small LINE RNAs may represent precursors of the piRNAs.
Figure 7. Expression of small RNAs from P1-LINE in adult rat tissues. Expression of small RNA from P1-LINE was examined in eight different tissues of the adult rat by denaturing (12% polyacrylamide+8M urea) gel electrophoresis followed by northern blot hybridization. (A) An ethidium bromide-stained gel of the rat liver RNA shows the 5.8S rRNA (163 nt.), 5S rRNA (120 nt.) and tRNA (75 nt.). (B) Hybridization of the similar gel with 32P-labeled 310 bp P1-LINE DNA probe. Small RNAs of ~700–75 nt. are detected. M = molecular size marker (48–20 nt. oligos).
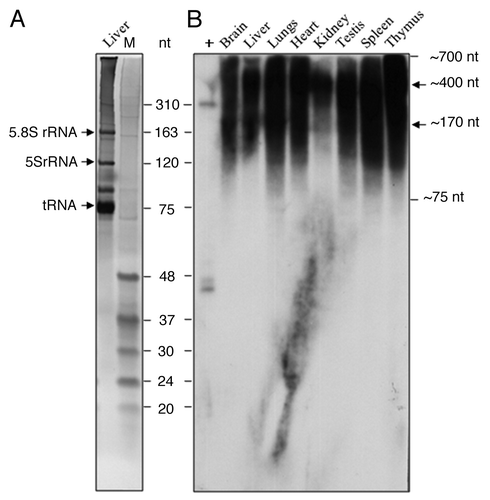
Table 5. Homology of P1-LINE with rat ESTs
Discussion
The present study has focused on endogenous RNA expression from LINEs in somatic tissues of the rat (Rattus norvegicus). We isolated a 2.8 kb PstI genomic repetitive DNA from the rat brain, which is a truncated LINE (L1) with homology to ORF2 (reverse transcriptase) of rat LINE 1 (RnL1).Citation14 P1-LINE is present as multiple 5′ truncated copies at different genomic loci in all rat chromosomes. These P1-LINE sequences are hypermethylated and show tissue-specific, mosaic distribution in rat genome. P1-LINE shows widespread RNA expression in somatic tissues in a heterogeneous, tissue-specific, dynamic and methylation-unrestricted manner. Most of these LINE RNAs from variously truncated LINEs represent long noncoding RNAs (lncRNAs) and precursors for piRNAs arguing for the noncoding RNA based cellular function(s) of LINEs.
Organization of P1-LINE sequences in the rat genome
We isolated, cloned and sequenced six repetitive DNA fragments (B1, E1, E2, H1, P1 and P2) from genomic DNA of the rat brain (; ; Fig. S1). DNA sequence analysis showed that out of the six DNA fragments, P1, E1 and H1 contain 92%, 80% and 43% LINE sequences, respectively. P1-LINE sequences are present in multiple copies at different positions in all rat chromosomes (; ) representing LINE sequences of the rat genome. The 123–2796 bp region of P1-LINE is 92% homologous to the rat L1 sequence (RnL1), but it is discontinuous and split into three parts (123–1162 bp, 1231–1404 bp and 1424–2796 bp) by the simple repeat and intergenic sequences (). P1-LINE sequence homologous to ORF2 is present in the 3′end (4164–5859 bp) of the Rn L1. A total of 2116 bp (123–1162 bp+1231–1404 bp+1424–2796 bp) of P1-LINE is homologous to 1695 bp (4164–5859 bp) of ORF2 of RnL1; and 447 bp sequence of RnL1 ORF2 (4962–5408 bp) is duplicated in P1-LINE, i.e., at 123–562 bp and 2347–2794 bp. These two regions of P1-LINE are 94% and 82% similar to ORF2, respectively. Initially, they might have been part of two different L1 sequences and during the course of evolution they might have joined together.Citation46 The discontinuity or split in the ORF2 sequences of P1-LINE also supports this idea. LINE sequences are generally present in the AT-rich regions of mammalian genome.Citation47 The 2.8 kb P1-LINE is present in the 93 kb sub-telomeric region of chromosome 12 (12p12) (), which is also AT-rich and devoid of exonic sequences. The 1400–2794 bp region of P1-LINE (94% homologous to ORF2 of RnL1) is named as common region (P1-LINEc). Interestingly, P1-LINEc is present in multiple copies at number of loci in all rat chromosomes representing its widespread distribution in the genome (; ). These LINE sequences are 5′ truncated (devoid of 5′-UTR promoter) and possess mutations in the ORF2 and therefore should be defective for retrotransposition. This agrees to the idea that the 3′end of L1 sequences are widely distributed and move frequently in the genome during evolution.Citation11 The fact that LINE sequences are normally present in clusters in the genome, is supported by the 93 kb region of chromosome 12 (12p12) containing P1-LINE, 52 truncated LINEs and two full length L1s (). This full length L1 is capable of making ORF1 protein but not ORF2 protein, as it contains several mutations as translational termination sites. The 93 kb region also contains 57 SINEs along with the LINEs showing its dependence on L1 for retrotransposition and thus, its non-autonomous nature.Citation48 It would be interesting to study the chromatin structure of this LINE-SINE cluster in the subtelomeric 93 kb (12p12) region of the rat chromosome 12 and its relationship to the LINE/SINE RNA expression.
Widespread expression of P1-LINE as long noncoding RNAs in somatic tissues
We observed that three genomic repetitive DNA fragments, i.e., P1, E1, H1 with LINE sequences are expressed as endogenous RNAs in the rat tissues (; Fig. S2; and ). Other three genomic repetitive DNA fragments, i.e., B1, E2, P2, which contain far less %age or no LINE sequence, are either weakly expressed or not expressed into RNA in the rat tissues (Fig. S2). Earlier, germ-line expression of L1 sequences has been reported in mouse testis and mouse ovary and human testis but there are very few reports of expression of LINEs in somatic cells or tissues of adult mammals.Citation49-Citation52 We observed widespread expression of P1-LINE RNAs as long noncoding RNAs in seven different somatic tissues (brain, liver, lungs, heart, kidney, spleen and thymus) and one germline tissue (testis) of adult (16 week) () and immature (4 week) rats (Fig. S3A). The expression of LINE RNAs is heterogeneous, tissue-specific and variable in individual rats suggesting its transcription from truncated LINEs at different chromosomal loci and its dynamic nature of expression in the genome. Northern blot analysis showed larger transcripts of ~5.0 > 2.8 kb and smaller transcripts of < 2.8 kb suggesting transcription from the truncated copies of P1-LINE. Individual rats showing variable RNA expression patterns with respect to the tissues suggest dynamic expression of the LINEs. Since, 28S and 18S rRNAs may contain certain LINE sequences, we also observed some P1-LINE RNA signals similar to the sizes of these rRNAs.Citation53,Citation54 We observed more LINE RNAs in the range of 1.5 kb to 0.2 kb in the northern blots (; Fig. S2), which correlate with genomic distribution of the LINEs (), shows widely distributed P1-LINE common region (1400–2796 bp) in rat genome. This shows that multiple P1-LINE copies located at different positions in the chromosomes are transcribed to generate the heterogeneous and dynamic long noncoding LINE RNA expression pattern. Northern blots (; Fig. S2) show that P1-LINE RNAs are expressed in the heart, kidney, testis, spleen, thymus, brain, lungs and liver from high to low levels with variations in the level of expression and transcript size in individual rats. RT-PCR () shows that P1-LINE RNAs are expressed in brain, testes, thymus, spleen, lungs, kidney, liver and heart from high to low levels. This difference in the expression patterns observed by the two techniques should be due to variations in the presence of truncated P1-LINE copies and their transcriptional activity in the tissues. This suggests transcription of P1-LINE being regulated in a tissue-specific and dynamic manner. The immature (4 week) and adult (16 week) rats both show P1-LINE RNA expression suggesting the onset of LINE transcription from early part of life in mammals. Previously expression of LINEs has been reported to occur under cellular stress conditions, e.g., oxidative stress in human neuroblastoma cells, gamma radiation in osteosarcoma cells, induction by a tryptophan-photoproduct in HeLa cells and induction by benzo(a)pyrene in HeLa cells.Citation55-Citation58 Also, cancerous cells and tissues have been reported to show higher expression of LINEs, e.g., colon cancer, renal cell carcinoma, prostate cancer, liver carcinoma, gastric cancer, ovarian carcinoma and teratocarcinoma as well as in other diseases such as hemophilia, Duchenne Muscular Dystrophy (DMD), β-thalassemia, chronic granulomatous disease (CGD) and Coffin-Lowry Syndrome (CLS).Citation59-Citation71 Therefore, expression of L1s is generally correlated with genome rearrangement, genomic instability, alterations in gene expression and diseases.Citation72 Recently, LINEs have been reviewed as a major source of genetic disposition to diseases in humans.Citation73-Citation75 However, this is the first report of endogenous LINE RNA expression as long and small noncoding RNAs in seven different somatic tissues and one germline tissue of a mammalian species under normal physiological conditions. This argues for biological role of LINE RNAs as cellular noncoding RNAs. These LINE RNAs may be associated with the chromatin and involved in organizing chromatin domains/landscapes for regulation of gene expression, e.g., LINE RNA associated with the Xist noncoding RNA for inactivation of mammalian X chromosome or acting as long noncoding RNAs for binding with regulatory proteins, e.g., the hsrωRNA during the heat shock response in Drosophila.Citation76,Citation77
P1-LINE DNA methylation and RNA expression
LINE sequences have self-limiting properties toward retrotransposition, e.g., deletion of promoter in the 5′-UTR and mutation in the ORF2, besides active silencing mechanisms, such as, epigenetic regulations by DNA methylation, histone methylation, small regulatory RNA-mediated heterochromatinization and inhibition by APOBEC3 (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide -a family of DNA or RNA cytidine deaminases).Citation7,Citation78-Citation82 It also includes AT-richness of L1 causing inefficient transcription and premature polyadenylation to inhibit its expansion.Citation10,Citation83 Methylation at CpG-dinucleotides in the GC-rich promoter of L1 causes silencing by inhibiting the initiation of transcription.Citation84 However, cytosine methylation has been reported to be preferentially directed to genes not to retrotransposons in a urochordate.Citation85 Similarly, in another example, methylation status of the LTR-retrotransposon did not correlate with the copy numbers and activity.Citation86
Our results show that P1-LINE is strongly hypermethylated in all the eight tissues studied, while a few of them like heart and testis show some degree of hypomethylation (). The hypermethylation status of P1-LINE does not correlate with its strong RNA expression in the tissues, which cannot be accounted by the small fraction of the hypomethylated P1-LINE sequences. We also observed tissue-specific methylation patterns of P1-LINE with respect to the hypermethylated 5kb Msp I band in the lungs and thymus and the hypomethylated 2.8 kb, 1.3 kb, 0.89 kb and 0.4 kb HpaII bands in the heart and testis suggesting variations in the organization of the LINE-chromatin in the tissues. Recent report shows that methylation within the genes (gene body methylation) is also important and involved in the transcriptional regulation.Citation87 Similarly, P1-LINE RNA expression may not be inhibited by DNA methylation. Since the strong, heterogeneous and widespread RNA expression suggests expression of LINEs from multiple sites in the rat genome, we consider this is unrestricted by the DNA hypermethylation. This observation was not reported earlier. It would be interesting to study the methylation status of P1-LINE chromatin in relation to its RNA expression and relationship of long noncoding LINE RNA with DNA methylation.
P1-LINE shows tissue-specific mosaic pattern in rat genome
Due to high copy number and widespread presence of LINEs in the genome, it gives mosaic appearance.Citation88 IRS-PCR patterns of P1-LINE by single primer () show that different copies of P1-LINE are present as inverted repeats in close proximity of each other in the rat genome. Different sizes and intensities of the DNA-bands amplified by a single primer (P1 or P2) represent multiple copies of LINEs as inverted repeats at different genomic locations. The differential pattern of IRS-PCR for the eight different tissues clearly shows that the chromosomal organization of P1-LINE is unique in each tissue creating a tissue-specific and mosaic pattern of the genome (). Since, P1-LINE is strongly expressed at RNA level from multiple loci, its mosaic distribution and transcription together may create tissue-specific noncoding LINE RNA-landscapes of the chromatin of the genome. The hypermethylated state of P1-LINE may also add new property to this landscaping of the genome.Citation89 Earlier, it was shown that human populations contain about 44% polymorphic L1s in the genome. This accounts for 6 major classes of L1s (hot L1s) and an average human being may contain about 80–100 retrotransposition-competent L1s.Citation90 RNA expression and retrotransposition of L1 have been shown to contribute to mosaic cellular patterns in the mammalian brain and this has been reported to be involved in the process of neurogenesis in the adult brain, which is induced by cellular signaling mechanisms.Citation91,Citation92 Interestingly, LINE 1 retrotransposition has also been linked to excercise and experience leading to learning and memory in the hippocampus region of the mouse brain suggesting their physiological significance.Citation93 Somatic mosaicism and copy number variation of DNA sequences in the human tissues have also been reported to contribute to variations in the genotype as well as the phenotype of the tissues.Citation94,Citation95 Our results show that LINEs have a natural and tissue-specific mosaic genomic distribution in mammals.
P1-LINE and small RNAs
Recently, regulation of LINEs and other retrotransposons by piRNAs has been reported.Citation40,Citation96,Citation97 Presence of bidirectional promoters in the L1 sequences are involved in generation of the long double-stranded transcripts, which can act as precursors for the piRNAs. Sequence analysis of P1-LINE showed five rat piRNAs, which can be generated from the common region (1706–2513 bp) of P1-LINE present all over the genome. The high copy number of P1-LINE in the rat chromosomes () also correlates with the high copy number of the five piRNAs () produced from the rat genome. We observed strong expression of small transcripts of 700–75 nt homologous to the 310 bp probe derived from the P1-LINE common region (ORF2 and piRNA sequences) in the denaturing northern blot analysis of RNA from eight different tissues of the adult (16 week) () and immature (4 week) (Fig. S3B) rats. Moreover, three piRNAs (piR3, piR4, piR5) out of the five piRNAs are mapped to the 310 bp probe, which detected the small transcripts. Seven ESTs of 474–699 nt from the rat genome () also show strong homology with P1-LINE common region (1424–2796 bp). This suggests that the small P1-LINE transcripts (700–75 nt) are partly homologous to the 5′UTR and 3′UTR regions of the rat transcripts/mRNAs represented as the ESTs and they may also be precursor RNAs for the piRNAs. Interestingly, the P1-LINE common region is the ORF2 sequence. Hence, 5′truncated LINEs with ORF2 sequences abundantly present in the rat genome can produce precursor RNAs for the piRNAs. Since piRNAs can silence invasion of transposons into the genome, the transcripts from the truncated LINEs can negatively regulate retrotransposition of LINEs and may also regulate the transcripts/mRNAs either directly or through the piRNAs. This indicates biological role of the LINE RNAs as well as the piRNAs for RNA regulations in the somatic tissues of the rat. This is likely to be independent of the fate of the LINE RNAs for retrotransposition. Therefore, both these LINE RNAs and piRNAs should represent noncoding RNAs. Recently, LINE RNAs have been shown to contain self-cleaving catalytic RNAs or ribozymes for the 5′end-RNA processing of the LINE RNAs and translational initiation of the ORFs indicating a primary function for the RNA regulation.Citation98
Taken together, our results suggest that P1-LINE, abundantly present at multiple chromosomal sites in the rat genome, is transcriptionally highly active in almost all somatic and gametic tissues in the young and adult individuals. It shows tissue-specific and mosaic genomic distribution. P1-LINE DNA is heavily methylated in the chromatin, but it is strongly expressed into LINE RNAs. The truncated LINEs are expressed into heterogeneous long and small noncoding RNAs, which may mark RNA-landscapes in the chromatin and may also be precursors for piRNAs for small RNA-mediated cellular regulations. This is the first report of methylation-unrestricted, widespread, strong and dynamic flux of retrotransposon RNA expression from endogenous LINEs with mosaic genomic patterns in the somatic tissues of a mammalian species. Biological role of such LINE RNAs in mammalian tissues under normal physiological conditions should explain their functional significance in the context of noncoding RNA, genome expansion and evolution.
Materials and Methods
Animal, tissues and reagents
Healthy male immature (4 week) and adult (16 week) rats (Rattus norvegicus, Wistar strain) were obtained from the Animal House of Jawaharlal Nehru University, New Delhi. The study was performed as per the guidelines and approval of the Institutional Animal Ethics Committee (IAEC). Eight tissues (brain, liver, lungs, heart, kidney, testes, spleen and thymus) were collected and used for the experiments. Molecular biology grade reagents and oligonucleotides from Sigma-Aldrich (USA), restriction enzymes, DNA/RNA modifying enzymes and polymerases from New England Biolabs, Fermentas, Promega, α-(32P)dATP (specific activity = 4 × 106 CPM/mmole) from BRIT were used for the study. Basic molecular biology methods were followed as described earlier.Citation99
Genomic DNA isolation and DNA cloning
Genomic DNA (20 μg) purified from the rat brain was digested by BamH I, EcoR I, Hind III, PstI and the DNA fragments were resolved by 1% agarose-TAE buffer (40 mM TRIS-acetate, 1 mM EDTA, pH 8.0) gel electrophoresis with 0.5 μg/ml ethidium bromide. We observed two major PstI bands of approximately 2.8 kb (P1) and 2.3 kb (P2) in addition to the smear (20–0.5 kb). The P1 (2.8 kb) DNA fragment was cut out and extracted from the gel by using a gel-extraction kit (cat. no. 28704, Qiagen) and the DNA was cloned into the PstI site of pBS-SK (+) II vector. The plasmid DNA was isolated from the transformed colonies and the recombinant clones (pP1-LINE) were checked by PstI digestion for the 2.8 kb inserted DNA fragment. Other genomic DNA fragments (B1, E1, E2, H1 and P2; described in the results) were similarly purified, cloned and verified.
DNA sequencing and sequence analysis
The 2.8 kb PstI DNA clone was sequenced and analyzed in silico by homology search in BLAST (www.blast.ncbi.nlm.nih.gov), BLAT (www.genome.ucsc.edu), Repeat Masker (www.repeatmasker.org), Promoter 2.0 prediction (www.cbs.dtu.dk), PROMO 3.0 (www.alggen.lsi.upc.es) sites and by piRNA (PIWI-interacting RNA) database (pirnabank.ibab.ac.in). Homology searches with the rat genome (www.rgd.mcw.edu) and rat EST database (www.blast.ncbi.nlm.nih.gov) were also analyzed. For BLAT analyses, the selected hits with the score and identity of 1000 and 90% were considered, respectively. For the piRNAs, 100% identity was observed. The 2.8 kb sequence was also compared with the full length rat LINE 1 (RnL1, accession number DQ100473) reported earlier.Citation14 The B1, E1, E2, H1, P2 genomic DNA fragments were also sequenced and the sequences were analyzed in silico.
RNA isolation and northern blot analysis
Total RNA was isolated from the rat tissues (brain, liver, lungs, heart, kidney, testes, spleen and thymus) by the Tri-reagent (cat. no. T9424, Sigma) method. RNA expression was studied by northern blot hybridization. Fifteen μg of total RNA from each tissue was denatured by 50% formamide, 2.5 M formaldehyde in MOPS [3-(N-morpholino) propanesulfonic acid] buffer (20 mM MOPS, 2 mM sodium acetate, 1 mM EDTA, pH 7.0) at 65°C for 15 min, cooled, mixed with 2 × loading buffer [0.025% (w/v) bromophenol blue, 0.025% (w/v) xylene cyanol, 5 mM EDTA (pH 8.0)] and resolved by 1.5% agarose-formaldehyde gel electrophoresis in MOPS-buffer. The RNA was vacuum-blotted from the gel on to the nylon membrane (cat. no. 60106, Biodyne A) and immobilized by UV-cross-linking. The blot was stained with methylene blue to check for the quality and complete transfer of the RNA. The 2.8 kb P1-LINE DNA was released from the pP1-LINE clone by PstI-digestion, gel-purified and 32P-labeled by using [α-32P]dATP, random hexamer-primer and E. coli DNA Pol I (Klenow) by random-primed synthesis. The 32P-labeled DNA probe was purified by Sephadex G-50 spin-column. Pre-hybridization of the blot was performed in hybridization buffer {50% formamide, 5X SSC (20X SSC = 3M NaCl+0.3 M Na-citrate, pH 7.0), 0.05 M Na2HPO4, 0.5% SDS, 1 mM EDTA, 5X Denhardt’s reagent [0.1% (w/v) Ficoll, 0.1% (w/v) Polyvinylpyrrolidone and 0.1% (w/v) BSA], 100 μg/ml denatured carrier tRNA and 100 μg/ml denatured calf thymus DNA} at 42°C for 2 h. Hybridization of the blot was performed in the hybridization buffer added with the heat-denatured 32P-labeled DNA probe (specific activity = 2–3 × 108 cpm/μg) at 106 cpm/ml conc. at 42°C for 16–18 h. The membrane was washed in 5X SSC+0.1% SDS at 25–30°C for 10 min and successively in 2X, 1X, 0.1X SSC containing 0.1% SDS at 55°C once for 10 min each time to remove background and non-specific hybridization. The blot was exposed to X-ray film for autoradiography. Unlabelled P1-LINE DNA (100 pg) was used as a positive control. The 1kb denatured DNA-ladder on the membrane was stained with methylene blue and used to assess the size of the hybridization signal. A 416 bp DNA (5′1–416) from the rat 28S rRNA (accession # NR_046246.1) gene was PCR-amplified by the primers (forward primer: 5′CGCGACCTCAGATCAGACGTG3′ and reverse primer: 5′CACGCCCTCTTGAACTCTCT3′) from the rat genomic DNA, gel-purified, 32P-labeled and hybridized with the RNA gel blots after removing the P1-LINE DNA probe. The P1-LINE RNA and 28S rRNA signals were quantitated by densitometry as integrated density value (IDV) and the P1-LINE/28S rRNA normalized IDV ratio (Mean ± SEM) was plotted as relative RNA expression in the graph.
Reverse transcription-polymerase chain reaction (RT-PCR)
RNase-free DNase I-treated total RNA (1 μg) from the tissues was reverse-transcribed by reverse transcriptase (RT) and oligo(dT) primer in 20 μl reaction followed by PCR-amplification of 2 μl of the RT-reaction product by the primers [forward primer (P1): 5′TGTTGGGGCATCTTTTGGGTAT3′ and reverse primer (P2): 5′GGAATGTCGAATGGCTGAGAAA3′] derived from the 2421–2730 bp P1-LINE common (ORF2) region by using the following conditions: 94°C for 5 min and 25 times (94°C for 45 sec, 61°C for 45 sec, 72°C for 1 min) and extension at 72°C for 10 min. The 2.5 μl reaction product was resolved by 1.5% agarose-TBE buffer (45 mM Tris-borate, 1 mM EDTA, pH 8.3) gel electrophoresis. The -RT negative control and the GAPDH [primers: G1 (5′ACCACAGTCCATGCCATCAC3′) and G2 (5′TCCACCACCCTGTTGCTGTA3′), 452 bp amplicon] reference RT-PCR were performed. The 310 bp P1-LINE and the 452 bp GAPDH amplicons were quantitated by densitometry as IDV. The normalized ratio of the IDVs of the 310 bp/452 bp signals (Mean ± SEM) was plotted as graph.
DNA methylation by Southern blot analysis
Genomic DNA (30 μg) from the tissues was digested by PstI+Msp I and PstI+HpaII (10 Units/μg DNA, 16 h) and resolved by 1% agarose/TAE buffer gel electrophoresis. DNA was in-gel depurinated by 0.25 N HCl for 10 min, washed with H2O for 10 min, denatured by 1.5 M NaCl+0.5 M NaOH for 30 min (twice) and neutralized by 1.5 M NaCl + 1 M Tris-Cl (pH 7.0) for 30 min (twice). DNA was vacuum-blotted to the nylon membrane, UV-crosslinked, pre-hybridized in Sodium-Phosphate buffer (0.3 M Na2HPO4, 7% SDS, 1 mM EDTA and 100 μg/ml denatured calf thymus DNA) at 65°C for 2 h and hybridized in hybridization buffer added with denatured 32P-labeled 2.8 kb P1-LINE DNA probe (specific activity = 2–3 × 108 cpm/μg) at 106 cpm/ml conc. at 65°C for 16–18 h. The membrane was successively washed in washing solution I (1X SSC + 0.5% SDS) at 25–30°C for 10 min, washing solution II (0.6X SSC + 0.1% SDS) at 65°C for 20 min, washing solution III (0.2X SSC + 0.1% SDS) at 65°C for 20 min to remove background and non-specific hybridization. The membrane was exposed to X-ray film for autoradiography. Unlabelled 2.8 kb P1-LINE DNA (100 pg) was used as a positive control and P1-LINE DNA digested with Msp I or HpaII was loaded in the gel to locate the positions of the corresponding bands from the genomic DNA. Four distinct MspI-bands (2.8, 1.3, 0.89 and 0.4 kb) were quantitated by densitometry and their background-subtracted specific signal density (IDV) was used to calculate the % methylation as [(Msp I-HpaII)/Msp I] × 100 and plot the graph.
Interspersed repetitive sequence-polymerase chain reaction (IRS-PCR)
Genomic DNA purified from the tissues was used for Interspersed Repetitive Sequence-Polymerase Chain Reaction (IRS-PCR) by using the P1-LINE common region P1 and P2 primers (310 bp amplicon). The IRS-PCR (50 μl) mixture contained PCR-buffer (10 mM TRIS-HCl, 50 mM KCl, 2.5 mM MgCl2, pH 8.3), 50.0 ng genomic DNA, 0.2 mM dNTPs, 25.0 pmol of single primer (P1 or P2) or 12.5 pmol each of double primers (P1 + P2), 1.0 unit Taq DNA pol. As per our earlier report, the cycle parameters were as follows: denaturation at 94°C for 5 min; 10 cycles of 94°C for 50 sec, 30°C for 1 min 20 sec, 72°C for 3 min 30 sec; then 30 cycles of 94°C for 50 sec, 61°C for 1 min 20 sec, 72°C for 3 min 30 sec; and a final extension step at 72°C for 10 min; followed by an end-step at 4°C.Citation100 One-tenth (5 μl) of the PCR-product was analyzed by 1% agarose-TAE buffer gel electrophoresis. Another repeat-sequence DNA (accession number GQ463151), isolated from the rat genome, as well as the GAPDH gene were PCR-amplified from the genomic DNA by their corresponding specific primers for comparison. The IRS-PCR products (2/5th) were also mixed with alkaline-dye [50 mM NaOH, 1 mM EDTA, 3% Ficoll, 0.01% (w/v) bromophenol blue] and resolved in 1.5% alkaline-agarose gel in alkaline buffer (50 mM NaOH, 10 mM EDTA, pH 8.0) by electrophoresis at 3.5 V/cm, Southern-blotted to the membrane in the alkaline buffer and hybridized with 32P-labeled 310 bp P1-LINE common region DNA probe followed by washing of the blot and autoradiography.Citation101 The band patterns were scored. The hybridization signals were quantitated by densitometry and expressed as P1/P1+P2 and P2/P1+P2.
Denaturing (PAGE-urea) northern blot analysis for small RNA
Denaturing (PAGE-urea) northern blot hybridization was performed as described by Saito et al., (2006).Citation102 Briefly, 30 μg total RNA from the tissues was denatured in 50% formamide at 65°C for 15 min and mixed with 2× sample buffer [0.025% (w/v) bromophenol blue, 0.025% (w/v) xylene cyanol, 5 mM EDTA (pH 8.0)], resolved by 12% polyacrylamide-8M urea/TBE buffer denaturing gel electrophoresis and transferred to the nylon membrane by semi-dry electro-blotting in TBE buffer at 2 mAmp/cm2 for 90 min and immobilized by UV-crosslinking. Pre-hybridization of the blot was performed in hybridization buffer [5X Denhardt’s reagent, 5X SSC, 0.5% SDS, 1mM EDTA, 50 mM Na2HPO4, 100 μg/ml denatured calf thymus DNA and 100 μg/ml denatured carrier tRNA] for 2 h at 37°C. The 310 bp ORF2-probe (2421–2730 bp) was generated by PCR from P1-LINE DNA and 32P-labeled by random-primed synthesis as mentioned above. Hybridization of the blot was performed in the hybridization buffer added with the denatured 310 bp probe (specific activity = 2–3 × 108 cpm/μg) at 107 cpm/ml conc. at 37°C for 16–18 h. The membrane was washed twice with 2X SSC+0.1% SDS and once with 1X SSC+0.1% SDS at 37°C for 10 min each time to remove background and non-specific hybridization. The membrane was exposed to X-ray film for autoradiography. Unlabelled 310 bp ORF2-probe (100 pg) was used as a positive control.
The tissues from individual rats were analyzed to score variations in P1-LINE expression. RNA-expression, DNA-methylation and IRS-PCR analyses were performed in the tissues from the same individuals. Sizes of the bands were calculated from the standard curves of molecular size markers. The sizes of the RNAs calculated from the denatured DNA markers are mentioned as approximate values. The data were collected from 2–4 experiments and expressed as Mean ± SEM.
Additional material
Download Zip (792.5 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We acknowledge the Capacity-Buildup, University Grants Commission-Research Network Resource Centre, Department of Science and Technology-PURSE grants/facility to Jawaharlal Nehru University, School of Life Sciences and PCR as well as the Junior/Senior Research Fellowship from the Council of Scientific and Industrial Research to DKS from the Government of India.
Supplemental Materials
Supplemental materials may be found here: https://www.landesbioscience.com/journals/rnabiology/article/22402/
References
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al, International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 2001; 409:860 - 921; http://dx.doi.org/10.1038/35057062; PMID: 11237011
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, et al, Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature 2002; 420:520 - 62; http://dx.doi.org/10.1038/nature01262; PMID: 12466850
- Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, et al, Rat Genome Sequencing Project Consortium. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 2004; 428:493 - 521; http://dx.doi.org/10.1038/nature02426; PMID: 15057822
- Goodier JL, Kazazian HH Jr.. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 2008; 135:23 - 35; http://dx.doi.org/10.1016/j.cell.2008.09.022; PMID: 18854152
- Deininger PL, Batzer MA. Mammalian retroelements. Genome Res 2002; 12:1455 - 65; http://dx.doi.org/10.1101/gr.282402; PMID: 12368238
- Babushok DV, Kazazian HH Jr.. Progress in understanding the biology of the human mutagen LINE-1. Hum Mutat 2007; 28:527 - 39; http://dx.doi.org/10.1002/humu.20486; PMID: 17309057
- Ostertag EM, Kazazian HH Jr.. Biology of mammalian L1 retrotransposons. Annu Rev Genet 2001; 35:501 - 38; http://dx.doi.org/10.1146/annurev.genet.35.102401.091032; PMID: 11700292
- Beauregard A, Curcio MJ, Belfort M. The take and give between retrotransposable elements and their hosts. Annu Rev Genet 2008; 42:587 - 617; http://dx.doi.org/10.1146/annurev.genet.42.110807.091549; PMID: 18680436
- Kazazian HH Jr.. Mobile elements: drivers of genome evolution. Science 2004; 303:1626 - 32; http://dx.doi.org/10.1126/science.1089670; PMID: 15016989
- Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 2004; 429:268 - 74; http://dx.doi.org/10.1038/nature02536; PMID: 15152245
- Medstrand P, van de Lagemaat LN, Mager DL. Retroelement distributions in the human genome: variations associated with age and proximity to genes. Genome Res 2002; 12:1483 - 95; http://dx.doi.org/10.1101/gr.388902; PMID: 12368240
- Szak ST, Pickeral OK, Makalowski W, Boguski MS, Landsman D, Boeke JD. Molecular archeology of L1 insertions in the human genome. Genome Biol 2002; 3:h0052; http://dx.doi.org/10.1186/gb-2002-3-10-research0052; PMID: 12372140
- Bailey JA, Carrel L, Chakravarti A, Eichler EE. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc Natl Acad Sci U S A 2000; 97:6634 - 9; http://dx.doi.org/10.1073/pnas.97.12.6634; PMID: 10841562
- Kirilyuk A, Tolstonog GV, Damert A, Held U, Hahn S, Löwer R, et al. Functional endogenous LINE-1 retrotransposons are expressed and mobilized in rat chloroleukemia cells. Nucleic Acids Res 2008; 36:648 - 65; http://dx.doi.org/10.1093/nar/gkm1045; PMID: 18073200
- Kapranov P, St Laurent G. Dark Matter RNA: Existence, Function, and Controversy. Front Genet 2012; 3:60; http://dx.doi.org/10.3389/fgene.2012.00060; PMID: 22536205
- Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res 2012; 40:6391 - 400; http://dx.doi.org/10.1093/nar/gks296; PMID: 22492512
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 2010; 39:925 - 38; http://dx.doi.org/10.1016/j.molcel.2010.08.011; PMID: 20797886
- Ankö ML, Neugebauer KM. Long noncoding RNAs add another layer to pre-mRNA splicing regulation. Mol Cell 2010; 39:833 - 4; http://dx.doi.org/10.1016/j.molcel.2010.09.003; PMID: 20864030
- Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell 2011; 145:178 - 81; http://dx.doi.org/10.1016/j.cell.2011.03.014; PMID: 21496640
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011; 43:904 - 14; http://dx.doi.org/10.1016/j.molcel.2011.08.018; PMID: 21925379
- Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci 2006; 29:77 - 103; http://dx.doi.org/10.1146/annurev.neuro.29.051605.112839; PMID: 16776580
- Rossbach M. Non-Coding RNAs in Neural Networks, REST-Assured. Front Genet 2011; 2:8; http://dx.doi.org/10.3389/fgene.2011.00008; PMID: 22303307
- Mitra SA, Mitra AP, Triche TJ. A central role for long non-coding RNA in cancer. Front Genet 2012; 3:17; http://dx.doi.org/10.3389/fgene.2012.00017; PMID: 22363342
- Niland CN, Merry CR, Khalil AM. Emerging Roles for Long Non-Coding RNAs in Cancer and Neurological Disorders. Front Genet 2012; 3:25; http://dx.doi.org/10.3389/fgene.2012.00025; PMID: 22375145
- Mattick JS. RNA driving the epigenetic bus. EMBO J 2012; 31:515 - 6; http://dx.doi.org/10.1038/emboj.2011.479; PMID: 22293829
- Wang X, Song X, Glass CK, Rosenfeld MG. The long arm of long noncoding RNAs: roles as sensors regulating gene transcriptional programs. Cold Spring Harb Perspect Biol 2011; 3:a003756; http://dx.doi.org/10.1101/cshperspect.a003756; PMID: 20573714
- Derrien T, Guigó R, Johnson R. The Long Non-Coding RNAs: A New (P)layer in the “Dark Matter”. Front Genet 2011; 2:107; PMID: 22303401
- Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, et al, FANTOM Consortium, RIKEN Genome Exploration Research Group Phase I & II Team. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 2002; 420:563 - 73; http://dx.doi.org/10.1038/nature01266; PMID: 12466851
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al, FANTOM Consortium, RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group). The transcriptional landscape of the mammalian genome. Science 2005; 309:1559 - 63; http://dx.doi.org/10.1126/science.1112014; PMID: 16141072
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007; 316:1484 - 8; http://dx.doi.org/10.1126/science.1138341; PMID: 17510325
- Rinn JL, Euskirchen G, Bertone P, Martone R, Luscombe NM, Hartman S, et al. The transcriptional activity of human Chromosome 22. Genes Dev 2003; 17:529 - 40; http://dx.doi.org/10.1101/gad.1055203; PMID: 12600945
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, et al. Global identification of human transcribed sequences with genome tiling arrays. Science 2004; 306:2242 - 6; http://dx.doi.org/10.1126/science.1103388; PMID: 15539566
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 2005; 308:1149 - 54; http://dx.doi.org/10.1126/science.1108625; PMID: 15790807
- Khaitovich P, Kelso J, Franz H, Visagie J, Giger T, Joerchel S, et al. Functionality of intergenic transcription: an evolutionary comparison. PLoS Genet 2006; 2:e171; http://dx.doi.org/10.1371/journal.pgen.0020171; PMID: 17040132
- Nakaya HI, Amaral PP, Louro R, Lopes A, Fachel AA, Moreira YB, et al. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol 2007; 8:R43; http://dx.doi.org/10.1186/gb-2007-8-3-r43; PMID: 17386095
- Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, et al. The reality of pervasive transcription. PLoS Biol 2011; 9:e1000625 - , discussion e1001102; http://dx.doi.org/10.1371/journal.pbio.1000625; PMID: 21765801
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007; 128:1089 - 103; http://dx.doi.org/10.1016/j.cell.2007.01.043; PMID: 17346786
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 2007; 315:1587 - 90; http://dx.doi.org/10.1126/science.1140494; PMID: 17322028
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 2007; 129:69 - 82; http://dx.doi.org/10.1016/j.cell.2007.03.026; PMID: 17418787
- Halic M, Moazed D. Transposon silencing by piRNAs. Cell 2009; 138:1058 - 60; http://dx.doi.org/10.1016/j.cell.2009.08.030; PMID: 19766558
- Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 2007; 12:503 - 14; http://dx.doi.org/10.1016/j.devcel.2007.03.001; PMID: 17395546
- Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol 2009; 25:355 - 76; http://dx.doi.org/10.1146/annurev.cellbio.24.110707.175327; PMID: 19575643
- O’Donnell KA, Boeke JD. Mighty Piwis defend the germline against genome intruders. Cell 2007; 129:37 - 44; http://dx.doi.org/10.1016/j.cell.2007.03.028; PMID: 17418784
- Yan Z, Hu HY, Jiang X, Maierhofer V, Neb E, He L, et al. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res 2011; 39:6596 - 607; http://dx.doi.org/10.1093/nar/gkr298; PMID: 21546553
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, et al. Characterization of the piRNA complex from rat testes. Science 2006; 313:363 - 7; http://dx.doi.org/10.1126/science.1130164; PMID: 16778019
- Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res 2006; 16:78 - 87; http://dx.doi.org/10.1101/gr.4001406; PMID: 16344559
- Yang S, Smit AF, Schwartz S, Chiaromonte F, Roskin KM, Haussler D, et al. Patterns of insertions and their covariation with substitutions in the rat, mouse, and human genomes. Genome Res 2004; 14:517 - 27; http://dx.doi.org/10.1101/gr.1984404; PMID: 15059992
- Kroutter EN, Belancio VP, Wagstaff BJ, Roy-Engel AM. The RNA polymerase dictates ORF1 requirement and timing of LINE and SINE retrotransposition. PLoS Genet 2009; 5:e1000458; http://dx.doi.org/10.1371/journal.pgen.1000458; PMID: 19390602
- Branciforte D, Martin SL. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol Cell Biol 1994; 14:2584 - 92; http://dx.doi.org/10.1128/MCB.14.4.2584; PMID: 8139560
- Trelogan SA, Martin SL. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc Natl Acad Sci U S A 1995; 92:1520 - 4; http://dx.doi.org/10.1073/pnas.92.5.1520; PMID: 7878012
- Ergün S, Buschmann C, Heukeshoven J, Dammann K, Schnieders F, Lauke H, et al. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J Biol Chem 2004; 279:27753 - 63; http://dx.doi.org/10.1074/jbc.M312985200; PMID: 15056671
- Belancio VP, Roy-Engel AM, Pochampally RR, Deininger P. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res 2010; 38:3909 - 22; http://dx.doi.org/10.1093/nar/gkq132; PMID: 20215437
- Eickbush DG, Luan DD, Eickbush TH. Integration of Bombyx mori R2 sequences into the 28S ribosomal RNA genes of Drosophila melanogaster.. Mol Cell Biol 2000; 20:213 - 23; http://dx.doi.org/10.1128/MCB.20.1.213-223.2000; PMID: 10594024
- Ye J, Eickbush TH. Chromatin structure and transcription of the R1- and R2-inserted rRNA genes of Drosophila melanogaster.. Mol Cell Biol 2006; 26:8781 - 90; http://dx.doi.org/10.1128/MCB.01409-06; PMID: 17000772
- Giorgi G, Marcantonio P, Del Re B. LINE-1 retrotransposition in human neuroblastoma cells is affected by oxidative stress. Cell Tissue Res 2011; 346:383 - 91; http://dx.doi.org/10.1007/s00441-011-1289-0; PMID: 22160459
- Farkash EA, Kao GD, Horman SR, Prak ET. Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic Acids Res 2006; 34:1196 - 204; http://dx.doi.org/10.1093/nar/gkj522; PMID: 16507671
- Okudaira N, Iijima K, Koyama T, Minemoto Y, Kano S, Mimori A, et al. Induction of long interspersed nucleotide element-1 (L1) retrotransposition by 6-formylindolo[3,2-b]carbazole (FICZ), a tryptophan photoproduct. Proc Natl Acad Sci U S A 2010; 107:18487 - 92; http://dx.doi.org/10.1073/pnas.1001252107; PMID: 20852066
- Teneng I, Montoya-Durango DE, Quertermous JL, Lacy ME, Ramos KS. Reactivation of L1 retrotransposon by benzo(a)pyrene involves complex genetic and epigenetic regulation. Epigenetics 2011; 6:355 - 67; http://dx.doi.org/10.4161/epi.6.3.14282; PMID: 21150308
- Schulz WA. L1 retrotransposons in human cancers. J Biomed Biotechnol 2006; 2006:83672; http://dx.doi.org/10.1155/JBB/2006/83672; PMID: 16877821
- Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res 1992; 52:643 - 5; PMID: 1310068
- Florl AR, Löwer R, Schmitz-Dräger BJ, Schulz WA. DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Br J Cancer 1999; 80:1312 - 21; http://dx.doi.org/10.1038/sj.bjc.6690524; PMID: 10424731
- Florl AR, Steinhoff C, Müller M, Seifert HH, Hader C, Engers R, et al. Coordinate hypermethylation at specific genes in prostate carcinoma precedes LINE-1 hypomethylation. Br J Cancer 2004; 91:985 - 94; PMID: 15292941
- Kim MJ, White-Cross JA, Shen L, Issa JP, Rashid A. Hypomethylation of long interspersed nuclear element-1 in hepatocellular carcinomas. Mod Pathol 2009; 22:442 - 9; http://dx.doi.org/10.1038/modpathol.2008.203; PMID: 19136926
- Wang GS, Wang MW, Wu BY, Yang XY, Wang WH, You WD. LINE-1 family member GCRG123 gene is up-regulated in human gastric signet-ring cell carcinoma. World J Gastroenterol 2008; 14:758 - 63; http://dx.doi.org/10.3748/wjg.14.758; PMID: 18205268
- Dammann RH, Kirsch S, Schagdarsurengin U, Dansranjavin T, Gradhand E, Schmitt WD, et al. Frequent aberrant methylation of the imprinted IGF2/H19 locus and LINE1 hypomethylation in ovarian carcinoma. Int J Oncol 2010; 36:171 - 9; PMID: 19956846
- Skowronski J, Fanning TG, Singer MF. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol Cell Biol 1988; 8:1385 - 97; PMID: 2454389
- Kazazian HH Jr., Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 1988; 332:164 - 6; http://dx.doi.org/10.1038/332164a0; PMID: 2831458
- Narita N, Nishio H, Kitoh Y, Ishikawa Y, Ishikawa Y, Minami R, et al. Insertion of a 5′ truncated L1 element into the 3′ end of exon 44 of the dystrophin gene resulted in skipping of the exon during splicing in a case of Duchenne muscular dystrophy. J Clin Invest 1993; 91:1862 - 7; http://dx.doi.org/10.1172/JCI116402; PMID: 8387534
- Divoky V, Indrak K, Mrug M, Brabec V, Huisman THJ, Prchal JT. A novel mechanism of b thalassemia: the insertion of L1 retrotransposable element into b globin IVS II. Blood 1996; 88:148a
- Meischl C, Boer M, Ahlin A, Roos D. A new exon created by intronic insertion of a rearranged LINE-1 element as the cause of chronic granulomatous disease. Eur J Hum Genet 2000; 8:697 - 703; http://dx.doi.org/10.1038/sj.ejhg.5200523; PMID: 10980575
- Martínez-Garay I, Ballesta MJ, Oltra S, Orellana C, Palomeque A, Moltó MD, et al. Intronic L1 insertion and F268S, novel mutations in RPS6KA3 (RSK2) causing Coffin-Lowry syndrome. Clin Genet 2003; 64:491 - 6; http://dx.doi.org/10.1046/j.1399-0004.2003.00166.x; PMID: 14986828
- Hancks DC, Kazazian HH Jr.. Active human retrotransposons: variation and disease. Curr Opin Genet Dev 2012; 22:191 - 203; http://dx.doi.org/10.1016/j.gde.2012.02.006; PMID: 22406018
- Belancio VP, Deininger PL, Roy-Engel AM. LINE dancing in the human genome: transposable elements and disease. Genome Med 2009; 1:97; http://dx.doi.org/10.1186/gm97; PMID: 19863772
- Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet 2011; 12:187 - 215; http://dx.doi.org/10.1146/annurev-genom-082509-141802; PMID: 21801021
- Solyom S, Kazazian HH Jr.. Mobile elements in the human genome: implications for disease. Genome Med 2012; 4:12; http://dx.doi.org/10.1186/gm311; PMID: 22364178
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell 2010; 141:956 - 69; http://dx.doi.org/10.1016/j.cell.2010.04.042; PMID: 20550932
- Prasanth KV, Rajendra TK, Lal AK, Lakhotia SC. Omega speckles - a novel class of nuclear speckles containing hnRNPs associated with noncoding hsr-omega RNA in Drosophila. J Cell Sci 2000; 113:3485 - 97; PMID: 10984439
- Yu F, Zingler N, Schumann G, Strätling WH. Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic Acids Res 2001; 29:4493 - 501; http://dx.doi.org/10.1093/nar/29.21.4493; PMID: 11691937
- Soifer HS, Zaragoza A, Peyvan M, Behlke MA, Rossi JJ. A potential role for RNA interference in controlling the activity of the human LINE-1 retrotransposon. Nucleic Acids Res 2005; 33:846 - 56; http://dx.doi.org/10.1093/nar/gki223; PMID: 15701756
- Yang N, Kazazian HH Jr.. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol 2006; 13:763 - 71; http://dx.doi.org/10.1038/nsmb1141; PMID: 16936727
- Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O’Shea KS, Moran JV, et al. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A 2006; 103:8780 - 5; http://dx.doi.org/10.1073/pnas.0603313103; PMID: 16728505
- Muckenfuss H, Hamdorf M, Held U, Perkovic M, Löwer J, Cichutek K, et al. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem 2006; 281:22161 - 72; http://dx.doi.org/10.1074/jbc.M601716200; PMID: 16735504
- Perepelitsa-Belancio V, Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat Genet 2003; 35:363 - 6; http://dx.doi.org/10.1038/ng1269; PMID: 14625551
- Hata K, Sakaki Y. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene 1997; 189:227 - 34; http://dx.doi.org/10.1016/S0378-1119(96)00856-6; PMID: 9168132
- Simmen MW, Leitgeb S, Charlton J, Jones SJ, Harris BR, Clark VH, et al. Nonmethylated transposable elements and methylated genes in a chordate genome. Science 1999; 283:1164 - 7; http://dx.doi.org/10.1126/science.283.5405.1164; PMID: 10024242
- Nakayashiki H, Ikeda K, Hashimoto Y, Tosa Y, Mayama S. Methylation is not the main force repressing the retrotransposon MAGGY in Magnaporthe grisea. Nucleic Acids Res 2001; 29:1278 - 84; http://dx.doi.org/10.1093/nar/29.6.1278; PMID: 11238993
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 2010; 328:916 - 9; http://dx.doi.org/10.1126/science.1186366; PMID: 20395474
- Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev 2009; 23:1303 - 12; http://dx.doi.org/10.1101/gad.1803909; PMID: 19487571
- Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature 2011; 479:534 - 7; http://dx.doi.org/10.1038/nature10531; PMID: 22037309
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A 2003; 100:5280 - 5; http://dx.doi.org/10.1073/pnas.0831042100; PMID: 12682288
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 2005; 435:903 - 10; http://dx.doi.org/10.1038/nature03663; PMID: 15959507
- Kuwabara T, Hsieh J, Muotri A, Yeo G, Warashina M, Lie DC, et al. Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat Neurosci 2009; 12:1097 - 105; http://dx.doi.org/10.1038/nn.2360; PMID: 19701198
- Muotri AR, Zhao C, Marchetto MC, Gage FH. Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus 2009; 19:1002 - 7; http://dx.doi.org/10.1002/hipo.20564; PMID: 19771587
- Piotrowski A, Bruder CE, Andersson R, Diaz de Ståhl T, Menzel U, Sandgren J, et al. Somatic mosaicism for copy number variation in differentiated human tissues. Hum Mutat 2008; 29:1118 - 24; http://dx.doi.org/10.1002/humu.20815; PMID: 18570184
- De S. Somatic mosaicism in healthy human tissues. Trends Genet 2011; 27:217 - 23; http://dx.doi.org/10.1016/j.tig.2011.03.002; PMID: 21496937
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell 2009; 136:656 - 68; http://dx.doi.org/10.1016/j.cell.2009.01.045; PMID: 19239887
- Lukic S, Chen K. Human piRNAs are under selection in Africans and repress transposable elements. Mol Biol Evol 2011; 28:3061 - 7; http://dx.doi.org/10.1093/molbev/msr141; PMID: 21613236
- Ruminski DJ, Webb CH, Riccitelli NJ, Lupták A. Processing and translation initiation of non-long terminal repeat retrotransposons by hepatitis delta virus (HDV)-like self-cleaving ribozymes. J Biol Chem 2011; 286:41286 - 95; http://dx.doi.org/10.1074/jbc.M111.297283; PMID: 21994949
- Sambrook J, Russell DW. Molecular Cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, New York, 2001.
- Dey I, Rath PC. Correlation between alterations in nucleosomal organization of LINEs in the promoter of cytochrome P450 2B1/2 gene and induction of CYP 2B1/2B2 mRNA expression by phenobarbitone in rat liver. DNA Cell Biol 2005; 24:359 - 70; http://dx.doi.org/10.1089/dna.2005.24.359; PMID: 15941388
- McDonell MW, Simon MN, Studier FW. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol 1977; 110:119 - 46; http://dx.doi.org/10.1016/S0022-2836(77)80102-2; PMID: 845942
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 2006; 20:2214 - 22; http://dx.doi.org/10.1101/gad.1454806; PMID: 16882972