Abstract
RNA folding is an essential aspect underlying RNA-mediated cellular processes. Many RNAs, including large, multi-domain ribozymes, are capable of folding to the native, functional state without assistance of a protein cofactor in vitro. In the cell, trans-acting factors, such as proteins, are however known to modulate the structure and thus the fate of an RNA. DEAD-box proteins, including Mss116p, were recently found to assist folding of group I and group II introns in vitro and in vivo. The underlying mechanism(s) have been studied extensively to explore the contribution of ATP hydrolysis and duplex unwinding in helicase-stimulated intron splicing. Here we summarize the ongoing efforts to understand the novel role of DEAD-box proteins in RNA folding.
Introduction
Group I and group II introns are both large, multi-domain RNAs that are capable of self-splicing. Like most RNAs, these introns have to adopt a specific 3D architecture to be functional.Citation1-Citation12 In vitro, most members of these two classes of introns were found to depend on non-physiological ionic concentrations and elevated temperature for optimal folding and in turn splicing.Citation13-Citation22 In the cell group I and II introns, however, require protein cofactors, such as intron-encoded maturases or host-encoded splicing factors, to promote their own excision from the precursor RNA.Citation15,Citation23-Citation29 Among these splicing factors is the nuclear-encoded protein Mss116p, a DEAD-box helicase, which is essential for splicing of all yeast mitochondrial group I and II introns.Citation30-Citation32 This raises the question of how these proteins shape the folding landscape of introns.
RNA typically encounters two significant folding problemsCitation33: i) Despite its apparent simplicity, RNA is prone to misfolding, resulting in inactive, often long-lived conformations, whereby the escape from the trapped state is the rate-limiting step in folding. ii) On the other hand, the native, functional RNA conformation or on-pathway intermediate structures are not necessarily thermodynamically stable and favored over other folding intermediates. Different proteins have been described to be capable of assisting RNA in overcoming their folding problem. For example, specific RNA binding proteins are required for stabilization of the RNA tertiary structure and in turn to overcome the thermodynamic barrier of productive RNA folding.Citation15,Citation23,Citation27,Citation29,Citation34-Citation45 These proteins were found to recognize and bind a distinct sequence or structure, thereby guiding folding and stabilizing the structure of their target RNA.Citation15,Citation23,Citation27,Citation29,Citation34-Citation45 In contrast, non-native interactions can be as stable as native contacts, thus misfolded structures are commonly long-lived (min to hours), trapping the RNA in an off-pathway intermediate.Citation13,Citation15,Citation16,Citation18-Citation22 The escape from such a kinetic trap is facilitated by RNA chaperones.Citation13,Citation15,Citation16,Citation18-Citation22 Transient binding of proteins with RNA chaperone activity destabilizes misfolded conformations, enabling the transition between on- and off-pathway intermediates (compare ref. Citation22). Recently, a number of RNA helicases have also been implicated in stimulating RNA folding.Citation29,Citation46 Interestingly, one member of the RNA helicase family, Mss116p, has so far been found capable of assisting RNA in overcoming both thermodynamic and kinetic barriers of folding.Citation47-Citation62
DEAD-box helicases are a ubiquitous protein family found in all kingdoms of life and involved in virtually all aspects of RNA metabolism, ranging from transcription, splicing and translation to mRNA decay.Citation63-Citation69 These enzymes show a strict preference toward RNA as substrate and hydrolyze ATP only.Citation63-Citation69 They contribute to cellular RNA-dependent processes by promoting RNA structural rearrangements and RNP remodeling in an ATP-dependent manner.Citation63-Citation69 In contrast to other SF2 helicases (e.g., NS3A and NphII), members of the DEAD-box helicase subfamily are non-processive.Citation63-Citation69 Interestingly, some DEAD-box helicases (e.g., Ded1p, Mss116p) are not only able to unwind RNA duplexes, but also to anneal RNA strands, whereby the latter activity is ATP-independent.Citation54,Citation60,Citation70 While most mechanistic insights were gained from studying helicases acting on model RNA substrates, the function of the yeast mitochondrial DEAD-box protein Mss116p has recently been examined for its cognate RNAs as well.Citation48-Citation58,Citation60,Citation62 Mss116p as well as the RNA helicases Ded1p from S. cerevisiae and Cyt19 from N. crassa have been shown to promote folding and in turn splicing of autocatalytic introns.Citation47-Citation62 The insights gained on the novel role of RNA helicases in facilitating RNA folding in vivo and at near-physiological in vitro are discussed in this review.
The Architecture of DEAD-Box Helicases
Like other helicases, DEAD-box proteins are composed of two globular domains that resemble the fold of the bacterial protein RecA involved in recombination.Citation64,Citation66-Citation68,Citation71 The covalently linked globular domains typically consist of a β-sheet surrounded by α-helices, whereby the number of β-strands and α-helices is to some extent variable. For example, the N-terminal RecA-like lobe of Mss116p contains an 8-stranded β-sheet bounded by 11 α-helices, while its C-terminal RecA domain consists of a 7-stranded β-sheet and 5 α-helices.Citation72 At least 11 characteristic sequence motifs are located at conserved positions within the two RecA-like domains (). While some of these motifs are found in the entire SF2 family, others are only conserved within the DEAD-box family.Citation63-Citation69 Notably, their name is derived from motif II, consisting of the amino acid sequence D-E-A-D. The N-terminal RecA-like domain (domain 1) harbors the ATP binding motifs Q and I, the ATP hydrolysis motifs II and III (SAT) and the RNA binding motifs Ia, Ib (GG) and Ic (TPGRxxD; ). The C-terminal RecA-like domain (domain 2) contains the RNA binding motifs IV, QxxR (IVa), V and Vb, the ATP binding motif VI and motif Va, which may coordinate ATPase and unwinding activities.Citation63-Citation69 In the absence of ATP and RNA the two helicase domains move freely with respect to each other (open conformation), but cooperative binding of ATP and RNA induces a compact helicase core structure (closed conformation), rationalizing the RNA-stimulated ATPase activity.Citation64,Citation66-Citation68,Citation71-Citation77 The two domains form a cleft into which the ATP cofactor binds.Citation64,Citation66-Citation68,Citation71 This ATP binding site is arranged in a highly defined manner, explaining the strong conservation of associated amino acids. In the crystal structures of a DEAD-box helicase bound to ssRNA,Citation72-Citation74,Citation76,Citation78,Citation79 the RNA was found to be situated opposite the ATP binding site and bound across both domainsCitation64,Citation66-Citation68,Citation71 (). In case of Vasa and Mss116p DEAD-box proteins, the helicase core establishes contacts to five or six nucleotides of the ssRNA molecule, respectively.Citation72,Citation76 Importantly, the conserved helicase motifs mostly contact the RNA sugar-phosphate backbone and several interactions are formed via the peptide backbone, explaining the lower sequence conservation within the RNA binding motifs. Importantly, the crystal structure of several DEAD-box proteins, including Vasa, Ddx19 and Mss116p,Citation72,Citation76,Citation78 revealed a bend within the RNA induced by motif Ic (TPGRxxD; formerly named motif 1bCitation64,Citation66,Citation80) in domain 1.Citation72-Citation76 As a consequence, the conformation, in which the ssRNA is bound to the helicase core, is incompatible with a helical RNA geometry. This kink may destabilize the RNA duplex as a first step toward unwindingCitation72,Citation76,Citation78; this however needs additional experimental evidence. Interestingly, in the case of Mss116p its helical CTE extends the RNA binding site and introduces a second bend in the bound RNACitation72 (). Truncation of the CTE severely reduces the binding affinity of Mss116p to intron RNAs (~10-fold or ~100-fold upon deletion of amino acids 569–664 or 551–664, respectively).Citation81 While ATP and RNA binding is well characterized, the intriguing question of how the ATP and RNA binding sites communicate is not fully understood. So far, motifs III and Va have primarily been implicated in the communication between nucleic acid and NTP binding sites.Citation82,Citation83 Mutations within these motifs impair coupling of ATP hydrolysis to RNA binding and in turn unwinding.
Figure 1. The DEAD-box helicase Mss116p and its conserved sequence motifs. Upper panel: Schematic representation of the helicase core region of the DEAD-box protein Mss116p. Mt, mitochondrial localization signal, which is cleaved off after import; NTE – N-terminal extension; CTE, C-terminal extension; C-tail, containing numerous basic amino acids. Functional regions shown in black or white, respectively, are not part of the 3D structure. Lower panel: Crystal structure of the Mss116p helicase core, which consists of two RecA-like domains, and its helical C-terminal extension. The conserved motifs are colored according to their primary function: red, ATP binding and hydrolysis; blue, RNA binding; yellow, communication between ATP binding and RNA binding sites. The non-conserved regions of the helicase domains are in gray and the CTE is shown in light-gray. The RNA is shown in pale yellow, the non-hydrolyzable ATP analog (AMPNP) in white and the Mg2+ ion in green. This figure has been adapted from references Citation68,Citation72,Citation80. Note: in some recent helicase reviews on DEAD-box helicases, motif Ib has been renamed to motif Ic, while the GG doublet has become motif Ib, thus we also refer to the GG doublet as motif Ib and to the TPGRLID sequence as motif Ic as described in references Citation64,Citation66,Citation80.
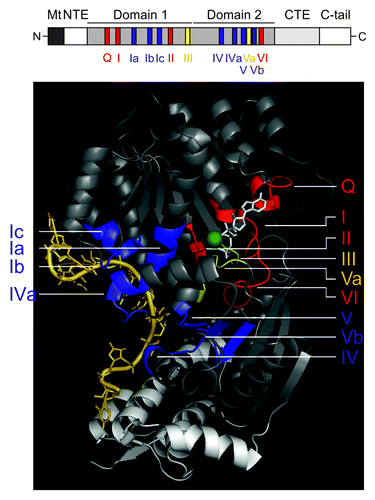
In all DEAD-box proteins, the helicase core is flanked by non-conserved C-terminal and N-terminal domains of variable sequence and length (few to hundreds of amino acids).Citation63-Citation69 In some cases, these terminal extensions harbor known motifs, such as RRMs, Zn-fingers and tudor domains, among others.Citation84 These auxiliary domains are thought to enable additional interactions with the target RNA, to recruit accessory proteins and to be critical for the physiological role of RNA helicases.Citation84 As most structural studies have been performed on truncated proteins, it is largely unknown how terminal domains are oriented with respect to the helicase core.Citation64,Citation66-Citation68,Citation71-Citation76 For example, in case of the RNA helicase YxiN, smFRET studies revealed that the RBD of YxiN lies on top of a slightly concave patch formed by flexible loops of the C-terminal RecA lobe.Citation85 Interestingly, the same patch is part of the interface between the DEAD-box protein eIF4A and its cofactor eIF4G,Citation86 whereas the extensions of Mss116p and Hera are oriented differently relative to the helicase core.Citation72,Citation87,Citation88
Mechanistic Insights into DEAD-Box Helicase Function
Although DEAD-box proteins are often part of large complexes, like the nascent ribosome or the spliceosome, mechanistic studies mainly focused on the RNA helicase working in isolation or in presence of a few cofactors. These structural and functional studies lead to a model in which DEAD-box proteins apply a mechanism of local duplex unwinding to achieve strand separation.Citation76-Citation78,Citation89 In this model, RNA helicases introduce a kink in the RNA backbone, thereby potentially destabilizing and locally unwinding the helixCitation76,Citation78; i.e., forcing the strands apart. However, there is evidence that kinking of the RNA is not sufficient for duplex unwinding but requires an additional step to complete the catalytic cycle.Citation77,Citation83 Applying an elegant smFRET approach Klostermeier and coworkers demonstrated that a conformational change is required but not sufficient for RNA unwinding by the RNA helicase YxiN.Citation83 While mutating the SAT motif of YxiN slowed down ATP hydrolysis and RNA unwinding, but did not affect the global structure of the close conformation, mutating the conserved glycin residue in motif Va prevents complete closure of inter-domain cleft, thereby affecting ATP binding and hydrolysis and being detrimental to unwinding.Citation83 Alternatively, DEAD-box proteins may simply trap the ssRNA strand and in turn inhibit duplex formation, thereby driving the overall reaction to the unwound state.Citation69
Even though the precise mechanism by which RNA helicases unwind duplexes remains elusive, it is evident that these proteins do not unwind helices by translocating along the RNA.Citation63-Citation69,Citation89-Citation92 This explains the fact that DEAD-box RNA helicases are only capable of unwinding short helices (10–12 bp up to two helical turns) in a non-processive manner and are sensitive to duplex stability.Citation46,Citation64,Citation68,Citation91,Citation93,Citation94 Notably, the helix length in cellular RNAs and RNPs rarely exceeds one helical turn. RNA helicases load directly onto the duplex and then separate the strands,Citation89,Citation92 explaining the lack of a strict unwinding polarity.Citation69 Single-stranded elements or even structured RNA flanking the duplex facilitate loading of the RNA helicase.Citation60,Citation61,Citation89,Citation92 Independent of the duplex length, a single ATP molecule is required per unwinding event.Citation95-Citation97 In contrast to ATP binding, ATP hydrolysis is not a pre-requisite for strand separation.Citation95,Citation96,Citation98,Citation99 Instead, ATP hydrolysis is necessary for the fast release of DEAD-box proteins from the RNA and thus for substrate turnover,Citation96,Citation97,Citation99,Citation100 as the affinity of DEAD-box helicases to RNA decreases strongly in the ADP-bound state.Citation77,Citation96,Citation101,Citation102 In fact, the YxiN helicase was shown to maintain a closed conformation during ATP hydrolysis.Citation98 Re-opening of the inter-domain cleft might be coupled to phosphate release and disrupts the bi-partite RNA binding interface and in turn triggers the release of the unwound RNA.Citation98,Citation100
In addition to unwinding, several DEAD-box proteins, like Mss116p and Ded1p, display strand annealing activity in vitro in an ATP-independent manner and are capable of dislodging a protein bound to RNA.Citation54,Citation67,Citation70,Citation103-Citation105 The annealing reaction is however not the reverse of unwindingCitation105 and protein displacement from RNA is independent of unwinding.Citation103,Citation104 These seemingly unrelated processes facilitate RNA and RNP remodeling of diverse complexes in cells. Essentially, annealing by RNA helicases may facilitate RNA folding, the intramolecular version of strand annealing,Citation106 while their unwinding mode appears to be well-adapted for local structural changes in assembled RNAs and RNPs.
A DEAD-Box Protein Stimulates Splicing of Mitochondrial Introns in Yeast
MSS116 was identified in a screen for nuclear mutants that are unable to grow by respiration on non-fermentable carbon sources when their mtDNAs contain introns.Citation31 Except for one group I intron, which is located in the large rRNA, these mitochondrial introns are all found in the coxI and cob genes. As expected from its core sequence, the crystal structure of Mss116p revealed that it adopts the typical fold of an RNA helicase (), whereby the C-terminal extension protruding from the second RecA-like lobe appears to be involved in RNA bindingCitation72 (). In line with its helicase fold, Mss116p displays unwinding and annealing in vitro and hydrolyzes ATP stimulated significantly by RNA.Citation54,Citation60,Citation89,Citation101 Most intriguing, however, was the fact that, like for stabilizing RNA cofactors, Mss116p was observed to reduce the [Mg2+] necessary for intron folding in vitro.Citation54,Citation60 To understand Mss116p’s function as splicing factor, the importance of its conserved motifs involved in ATP hydrolysis, RNA binding and unwinding has been dissected (; ).
Table 1. Effects of mutations on Mss116p function
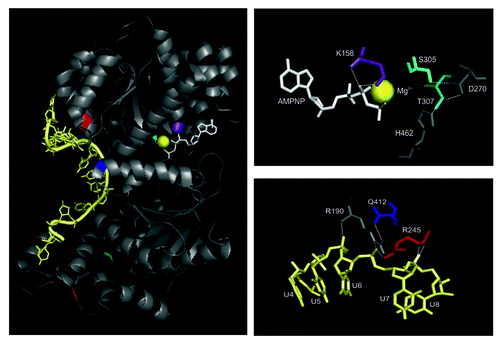
Figure 2. Interactions of the ATP analog and ssRNA with the helicase core. Left panel: Amino acids that were mutated to dissect Mss116p’s function in splicing of yeast mitochondrial introns are highlighted: K158, violet; S305, cyan; T307, dark cyan; Q412, blue; R245, red; I551, green; K569 – orange; the RNA is colored in pale yellow and AMPNP in white. The Mg2+ ion is represented as bright yellow sphere. Note that I551 and K569 are shown to indicate the start site of truncations in respective Mss116p mutants. Right panel: Conserved amino acids of the RecA-like domains contacting the non-hydrolyzable ATP analog (AMPNP; upper right panel) and RNA (lower right panel), respectively.
The role of ATP hydrolysis and unwinding by Mss116p in intron splicing
Mutating the conserved lysine (K158) in the Walker A motif revealed that ATP hydrolysis by Mss116p is required to promote intron splicing both in vitro and in vivo.Citation30,Citation49,Citation60 In contrast, it has been a matter of debate whether unwinding is also critical for this process.Citation49,Citation51,Citation58,Citation60,Citation62 Upon disrupting the extensive network of interactions critical for ATP binding and hydrolysis as well as unwinding, Caprara and coworkers recently revealed that the efficiency of ATP hydrolysis by Mss116p variants correlates with their ability to stimulate mitochondrial splicing; in contrast efficient unwinding is not sufficient for promoting intron splicingCitation49 (; ). For example, Q412A of the QxxR motif (IVa) disrupts the stabilizing inter-domain interaction with R190 of motif Ia, which directly interacts with the RNA near its bending point,Citation72 suggesting that this mutant might interfere with local strand separation. However, only a moderate effect on unwinding was observed (2.3-fold decrease in kobs).Citation49 Also, the growth on a non-fermentable carbon source (YP-Glycerol) and splicing of mitochondrial introns as well as ATP binding and hydrolysis appeared to be almost unaffected in the Q412A mutant.Citation49 In contrast, abolishing the direct contact between R245 in motif Ic (TPGRxxD; formerly named motif 1bCitation64,Citation66,Citation80) and RNA (near the kink)Citation72 strongly reduces ATP binding and hydrolysis (3–4 × effect on Km and kcat) and in turn growth on YP-Glycerol, splicing and duplex unwinding are strongly impaired in the R245E mutant.Citation49
Uncoupling ATP hydrolysis from RNA binding
The coupling energy between ATP and RNA binding was calculated to be 7.5 kJ/mol (1.8 kcal/mol) and 5.4 kJ/mol (1.3 kcal/mol) for the helicases YxiN and DbpA, respectively.Citation83,Citation107 Mutating the conserved glycine in motif Va (G303A in YxiN) or the SAT motif (S182A, T184A in YxiN), this coupling is reduced to ~3–4 kJ/mol (~0.7–1.0 kcal/mol).Citation83 Thus, such helicase variants prove useful in decoupling ATPase from unwinding activities. Like for other helicases, the SAT motif mutant of Mss116p (T307A) was found to bind ATP at a comparable level to wt Mss116p (Km of 180 ± 4 μM and 167 ± 20 μM for T307A and wt Mss116p, respectivelyCitation49), while its rate constant for ATP hydrolysis is 8-fold reduced (kcat of 3.66 ± 0.27 min−1 and 29.78 ± 3.62 min−1 for T307A and wt Mss116p, respectivelyCitation49). In line with the fact that solely ATP binding, but not ATP hydrolysis, is essential for duplex unwinding,Citation99 the T307A mutant displays only a 3.5-fold effect on the observed rate constant for unwinding.Citation49 However, the amplitude of unwinding is very small (< 20%).Citation49 This reduction in unwinding efficiency might be the result of a weakened RNA binding affinity, as observed for Ded1p.Citation82 As merely ATP hydrolysis is strongly affected in the T307A mutant, this appears to be the underlying cause for the observed growth inhibition.Citation49 Notably, the T307A mutation seems to strongly interfere with splicing in vivo in a yeast strain harboring all mt introns.Citation49 On the other hand, a S305A/T307A double mutant reduced splicing in vivo to ~50% in a strain devoid of all introns except ai5γ 58. As the S305A mutation does not interfere with Mss116p function in vivo, this implies that T307 is the central player in motif III.Citation49,Citation108,Citation109 Thus, these seemingly opposing results could indicate that the SAT motif and in turn unwinding plays a role in the coordination of the individual splicing events, as processing of the coxI and cob transcripts containing 7 or 5 introns, respectively, might dependent on temporal and structural regulation by a helicase. In vitro, the double mutant S305A/T307A, which binds ATP at wt level, is incapable of hydrolyzing ATP efficiently and barely unwinds RNA substrates in the size of a helical turn, which could be due to the somewhat reduced affinity for RNA.Citation51,Citation60 Nevertheless, this double mutant retains its ability to promote intron splicing in vitro,Citation60 which is consistent with the observation that Mss116p-promoted intron compaction is ATP-independentCitation53,Citation55; potentially the C-terminal domain makes up for the reduced RNA binding affinity and ATP hydrolysis-dependent protein recycling is less important during single turnover reaction conditions. These in vitro results on the S305A/T307A double mutant further lend support to the idea that the observed splicing inhibition in the T307A mutant strainCitation49 is related to functional timing of Mss116p. As T307 of motif III forms contacts with H462 of motif VI, involved in RNA binding, and with D270 of motif II, which is critical for ATP hydrolysis,Citation72 this mutant appears to effectively uncouple ATP binding and hydrolysis and in turn ATPase and unwinding activities. As Caprara and coworkers used a yeast strain harboring all mitochondrial introns, this suggests that both classes of self-splicing introns were affected to the same extent by the Mss116p mutants.Citation49
Genetic variation of Mss116p
In an high-throughput genetic screen Lambowitz and coworkers also identified conserved motifs associated with ATP and RNA binding or with inter-domain interactions as functionally important regions of Mss116p-stimluated splicing.Citation80 In addition, previously unidentified elements, which are located within surface loops, were implicated in Mss116p’s role as splicing factor and may function in protein-protein interactions.Citation80 Interestingly, several sequence variants of motif III, which couples ATP hydrolysis with RNA binding,Citation82 were identified in their genetic selections as well. This suggests that in vivo Mss116p tolerates a rather large number of sequence variations within the SAT motif and might be less dependent on this motif than other DEAD-box proteins due to the contribution of other protein domains to RNA binding.Citation80,Citation81,Citation110
Recycling of Mss116p
Since ATP hydrolysis is equally important for splicing of all yeast mt introns and releases Mss116p from its substrate RNA, this implies that recycling and turnover is absolutely critical for Mss116p function as a splicing factor and perhaps also for its role in RNA processing and translation in mitochondria.Citation49 As a consequence ATPase-deficient Mss116p variants might be sequestered on their target RNAs, thus limiting the available protein for participating in folding (and processing) of other RNA molecules. In turn, this “depletion” of the Mss116p pool might not only result in an increased population of thermodynamically unstable RNA intermediate, but also in a larger fraction of misfolded intron molecules and in turn an elevated need for unwinding activity of these mutant Mss116p variants. As Mss116p stimulates splicing of structurally distinct introns (requiring different additional protein cofactors) and supports mitochondrial RNA processing and translation, it has to be emphasized that it is difficult to assess the functional relevance of motifs associated with ATP binding and hydrolysis, unwinding as well as RNA binding in these distinct processes in vivo.
Stabilization of an Early Folding Intermediate along the Group II Intron Folding Pathway
In vitro, the yeast mitochondrial group II intron ai5γ follows a sophisticated folding pathway under non-physiological ionic concentrations and elevated temperature.Citation14,Citation111-Citation114 The ai5γ ribozyme first collapses slowly to a compact intermediate state followed by rapid progression to the native conformation.Citation111,Citation112 This slow collapse is controlled by the κ−ζ element, which is the anchor site for the catalytic center.Citation113,Citation114 As formation of an active site docking region is rate-limiting in vitro, this is an elegant way to ensure proper folding of the intron core and faithful splicing.Citation14,Citation112-Citation114 It is however known that in yeast mitochondria the RNA helicase Mss116p is essential for ai5γ splicing. In the past years, significant progress has been made in understanding how this DEAD-box protein facilitates intron folding.
Initially, mechanistic insights were inferred from the analysis of splicing kinetics of the ai5γ group II intron in the presence of Mss116p and mutant variants thereof. This lead to two models as to how Mss116p facilitates RNA folding: it might function as a splicing factor by acting in an RNA chaperone-like fashionCitation51,Citation54 or by providing stability to the intron RNA.Citation60 To obtain the first structure-based insights into Mss116p’s mechanism of action, Pyle and coworkers directly monitored DEAD-box protein-facilitated folding of the ai5γ intron in vitro.Citation53 Mss116p was found to directly stimulate ai5γ folding by accelerating the intron’s collapse to the near-native state through electrostatic stabilization of an early folding intermediate at near-physiological conditions in vitroCitation53 (). This work was further corroborated by a single molecule study on Mss116p-assisted ai5γ ribozyme folding by Rueda and coworkers.Citation55 Mss116p was observed to stimulate dynamic sampling between states along the ai5γ folding pathway, an effect previously observed only at high Mg2+ concentrations. Notably, facilitation of the intron collapse is independent of ATP, whereas ATP hydrolysis is required for the protein turnover.Citation96,Citation97,Citation99-Citation102 Importantly, Mss116p was not observed to stabilize the native state of the ai5γ intron, but stabilization comes from binding of flanking exon sequences.Citation53,Citation55
Figure 3. Mss116p-facilitated folding of the ai5γ intron in vitro. The DEAD-box protein accelerates compaction of intron domain D1 to a near-native folding intermediate, by stabilizing a specific structure at the core of this intron domain early in the ai5γ folding pathway. Subsequently, other intron domains can dock rapidly onto the D1 scaffold, whereby Mss116p does not stabilize the native state and is recycled upon ATP hydrolysis. D1 is shown in blue shades except for the κ−ζ element colored in purple shades, D3 is in green, D5 in red and D6 in yellow, while D2 and D4 are depicted in gray. Exons are outlined as thick black lines. Mss116p is indicated as orange ellipse. This figure has been adapted from references Citation52,Citation53.
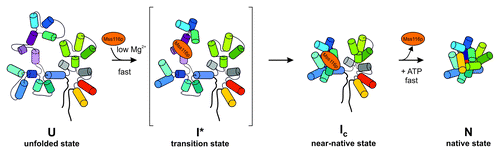
Most recently, an elegant, chemogenetic approach was employed to identify specific intron functional groups that participate in Mss116p-facilitated folding of an ai5γ splicing precursor.Citation52 In line with previous observations for the ai5γ ribozyme, in which the exons are deleted, compaction of the ai5γ pre-RNA is also limited by the κ−ζ substructure in D1, which serves as docking site for the active site center during later stages of intron folding.Citation52,Citation113,Citation114 Most importantly, the helicase was found to stabilize the κ−ζ element and in turn the early, obligate folding intermediate within intron domain 1, thereby laying the foundation for productive folding to the native state. In fact, nucleotide analogs that destabilize helices showed strong interferences in the presence of Mss116p, while analogs known to stabilize RNA helices displayed strong enhancements, suggesting that these atomic modifications interfere with or facilitate Mss116p-assisted ai5γ folding, respectively.Citation52 In addition, the exonic IBS2 region and the terminal intron sequence were implicated as important sites during folding of ai5γ pre-RNA.Citation52 This raises the question of whether long exons might interfere with correct structure formation at or close to the splice-sites.
Mss116p-Facilitated Ai5γ Folding in Yeast Mitochondria
Given the disparate environments during in vitro refolding and intracellular RNA structure formation,Citation29 a comprehensive comparison of RNA folding in vivo, and under non-physiological conditions in vitro is of tremendous interest. Therefore, we determined the intracellular structure of the ai5γ intron in different yeast genetic backgrounds using an in vivo structural probing technique.Citation56,Citation115 By monitoring Mss116p-induced conformational changes within the group II intron RNA in vivo, we gained the first detailed mechanistic insights into how this protein shapes folding of its target RNA in living cells. While the intron adopts the native conformation in the wt yeast strain, we found that ai5γ is able to form most of its secondary structure, but lacks its tertiary fold in the absence of Mss116p. Thus, the ai5γ intron appears largely unfolded in the mss116-knockout strain and depends on the DEAD-box protein at an early step of folding in vivo as well. Since the majority of the Mss116p-induced conformational changes reside within domain D1, Mss116p appears to assist in the formation of this largest domain, which is the scaffold for docking of other intron domains. These findings imply that Mss116p assists the ordered assembly of the ai5γ intron in vivoCitation56 and reveal interesting parallels between intracellular and in vitro Mss116p-promoted folding of a group II intron. So far, a stable misfolded structure has not been observed within the ai5γ intron in vivo,Citation56 but it cannot be ruled out yet whether the exons could interfere with correct folding of the splice sites.
Unwinding of Inhibitory Exon Structures and Non-Native Interactions by DEAD-Box Proteins
In general, the flanking sequence context is known to impact on RNA folding. For example, certain group I introns and RNase P RNA were found to become trapped in non-native conformations depending on the length and sequence of their flanking exons.Citation116-Citation125 In other words, studying RNA folding out of context might influence the folding pathway of an RNA. Indeed, while folding of the ai5γ ribozyme is devoid of kinetic traps,Citation14 abolishing the unwinding activity of Mss116p (mutant S305A/T307A) strongly affected splicing of a pre-RNA with long exons but not with short exons, indicating that unwinding is essential for exon unfolding, but not for intron folding.Citation62 Accordingly, Mss116p appears to play a dual role in intron folding in vitro, namely stabilization of the intron core and unwinding of inhibitory exonic elements trapping the pre-RNA in a splicing-incompetent conformation.Citation62,Citation80
Aside from non-native interactions involving the exons, it has been suggested that Mss116p assists in folding of ai5γ and other introns by unwinding misfolded secondary structure elements that interfere with assembling the native, splicing competent conformation.Citation50,Citation51,Citation54,Citation58 This model is supported by the observation that the DEAD-box proteins Cyt19 and Ded1p, which are able to functionally replace Mss116p in vivo and in vitro,Citation31,Citation35,Citation41,Citation102 destabilize non-native contacts in the Tetrahymena group I ribozyme.Citation47,Citation50 Recently, Russell and coworkers proposed that maximal folding of the ai5γ ribozyme and its pre-RNAs with either long or short flanking exons requires an ATP-dependent RNA unwinding by Mss116p.Citation58 This interpretation was based on the observation that the SAT mutant (S305A/T307A) promotes splicing of ai5γ less efficiently than the wild type Mss116p protein in vitro and in vivo and by the functional homology to Cyt19, which is capable of kinetically redistributing native and misfolded group I intron RNAs.Citation47 Importantly, if the reduced RNA binding affinity of this double mutant is taken into account,Citation60,Citation62 their results agree reasonably well with previously published data,Citation60,Citation62 which suggest that the unwinding deficiency is not the causal factor. Notably, Russell and coworkers obtained their results using a trans-cleavage system, in which the ai5γ ribozyme needs to bind and cleave a single-stranded substrate RNA. This complicates the interpretation of the data, because Mss116p binds ssRNA with high affinity,Citation54,Citation60 and it is likely to compete with the ribozyme for binding the substrate RNA. Of course, adding ATP to the reaction enhances protein turnover, thereby potentially increasing the fraction of substrate bound to the ribozyme. In brief, without structural evidence for misfolding within ai5γ (or any other yeast mt intron) and the action of Mss116p at the misfolded site, it remains possible that unwinding is not important for intron folding except for its role in resolving inhibitory structures at splice-sites and within exons.
How Does Mss116p Promote Folding of Structurally Distinct Intron RNAs?
The yeast mitochondrial genome encompasses both group I and group II introns, all of which depend on Mss116p for efficient splicing in vivo.Citation30 These two classes of autocatalytic RNAs do not share any sequence conservation and their structural organization is very different as well. Thus, it is of interest to understand whether this DEAD-box protein acts in a comparable manner on group II and group I introns. Since counterion-mediated compaction is an essential step in folding of all RNAs studied to date, this might argue for a conserved mechanism underlying the stimulation of intron splicing. However, splicing of the ai5γ intron is virtually abolished in an mss116-knockout strain, whereas group I introns display only a reduced splicing activity in the mss116-knockout strain and depend on additional protein cofactors for efficient splicing in vivo.Citation23-Citation27,Citation30,Citation35,Citation45,Citation48,Citation126-Citation131 For example, efficient bI5 intron splicing requires two nuclear-encoded proteins, namely Cbp2 and Mss116p, in vivo.Citation30,Citation31,Citation129 In a cbp2-knockout strain splicing of the bI5 intron is virtually abolished (< 1%), while only 40% of the cob pre-RNA are dependent on Mss116p, suggesting that Mss116p applies distinct mechanisms of action to promote folding of the ai5γ group II intron and the bI5 group I intron in vivo (Sachsenmaier and Waldsich, unpublished). In vitro Cbp2, which is a basic protein without any known motifs, chases the RNA from the collapsed state to the native state by capturing a compact, near-native bI5 folding intermediate.Citation44,Citation45,Citation131-Citation135 So far, the role of Mss116p in bI5 splicing remains enigmatic both in vitro and in vivo, but involves ATP-dependent steps.Citation49 Notably, as Cbp2 cannot discriminate between misfolded and correctly folded bI5,Citation136 this could explain the need for the DEAD-box helicase Mss116p; i.e., to resolve a trapped bI5-Cbp2-RNP complex.
Another yeast mitochondrial group I intron, named ai5β and located within the coxI pre-RNA, was found to be dependent on at least five nuclear-encoded proteins in vivo.Citation137 In vitro, however, two of these cofactors, Mrs1p and Mss116p, are sufficient to promote efficient ai5β splicing.Citation48 While the protein Mrs1p stabilizes the intron structure and assists in the first step of splicing, the RNA helicase Mss116p specifically enhances exon ligation in an ATP-dependent manner. Thus, Mss116p appears to utilize its ATPase function to coordinate the successive transesterification steps in splicing. It is also noteworthy that another DExH/D-box protein, Suv3p, is involved in promoting efficient ai5β intron splicing.Citation138 In Suv3p-deficient yeast strains stable, excised group I intron RNPs accumulate, thereby sequestering Mrs1p, which is crucial for splicing of both ai5β and bI3 mt group I introns, and in turn reducing splicing of aI5β. In brief, coordination of the individual steps in RNP assembly is likely to be crucial for efficient splicing of coxI and cob pre-RNAs, which harbor 7 and 5 of the 13 yeast mitochondrial introns, respectively, all of which depend on Mss116p. This raises the question of how “functional-timing” of DEAD-box protein action is achieved and whether it could be a limiting cofactor for splicing of some of these introns.
In case of the ai5β group I intron, Mss116p plays an entirely different role in promoting intron splicing compared with its function in ai5γ folding. However, it is not yet known whether a distinct mechanism is applied in stimulating splicing of other group I introns as well. Along this line, it also remains enigmatic whether Mss116p stabilizes early folding intermediates of other yeast mt group II introns.
Other DEAD-Box Proteins with a Basic C-terminal Extension and Their Role in RNA Folding
Aside from the conserved helicase motifs, Mss116p contains a helical C-terminal extension followed by an arginine-rich, positively charged C-tail. This C-terminal domain is shared by only a few other DEAD-box proteins, like Cyt19 and Ded1p.Citation81,Citation139 In fact, the N. crassa mitochondrial helicase Cyt19 as well as the S. cerevisiae non-mitochondrial protein Ded1p can functionally substitute for Mss116p, rescuing splicing of yeast mt group I and group II introns in an mss116-knockout strain.Citation30,Citation50,Citation54,Citation60 Studying the function of Ded1p on its natural target RNAs revealed that its primary function may not involve stimulation of RNA folding per se. Instead, this DEAD-box protein modulates translation by facilitating assembly and remodeling of the intermediate translation initiation complex eIF4F-mRNA.Citation140 Notably, the initial step in activating translation, in which Ded1p directly interacts with eIF4G to assemble a Ded1-mRNA-eIF4F complex, is ATP-dependent. A basic mechanistic framework for Ded1p-facilitated RNA structure conversion was provided by Jankowsky and coworkers.Citation105 On the one hand, Ded1p is able to disassemble RNA strands in an ATP-dependent manner, enabling the conversion of more stable into less stable RNA conformations. In the other pathway, Ded1p stabilizes intermediate structures in an ATP-independent manner. The latter activity could be responsible for Ded1p’s ability to promote intron folding by stabilizing an early folding intermediate along the ai5γ folding pathway in place of Mss116p and without requiring ATP hydrolysis.Citation99
Unlike Ded1p, the DEAD-box protein Cyt19 was identified to function in concert with Cyt18, a tyrosyl-tRNA synthetase, to promote group I intron splicing in vitro and in N. crassa mitochondria.Citation141 While Cyt18 guides intron folding by stabilizing on-pathway intermediates and preventing the formation of misfolded conformations,Citation36,Citation42,Citation43,Citation142 Cyt19, was proposed to destabilize non-native conformations along the Cyt18-assisted RNA-folding pathway in an ATP-dependent manner.Citation141 Interestingly, Cyt19 can act on several non-cognate intron RNAs, thereby stimulating their splicing and reverse splicing activities.Citation47,Citation57,Citation59,Citation61 Russell and coworkers demonstrated that Cyt19 mediates ATP-dependent unfolding of both the native conformation and a long-lived misfolded intermediate of the Tetrahymena group I intron.Citation47 The efficiency of unfolding was found to depend on the stabilities of the respective RNA species. Upon Cyt19-mediated unfolding, the RNA has a chance to refold and reach the splicing-competent state.Citation47 This observation is reminiscent of the way the RNA chaperone StpA resolves misfolded RNA structures.Citation42 In the same vein, both Cyt19 and Mss116p accelerate refolding of the Azoarcus group I intron in an ATP-dependent manner, presumably by resolving a misfolded, yet to be identified structural intermediate.Citation59 While the splicing stimulation of all these introns depends on ATP hydrolysis by Cyt19, it has not been determined whether discrete ATP-independent steps in promoting RNA folding precede the ATP-dependent folding transition, as it is the case for Mss116p in facilitating ai5γ folding.Citation52,Citation53,Citation55
Conclusion
RNA helicases have proven to be truly fascinating, versatile enzymes playing a role in nearly all aspects of RNA metabolism. With the discovery that DEAD-box proteins cannot only remodel RNA and RNP complexes, but are able to assist RNA in folding to its native conformation, a new, exciting aspect was added to RNA helicase function. In line with their action on model RNA substrates, which is their annealing and unwinding activity, some members of the DEAD-box protein family facilitate RNA folding by conferring stability to thermodynamically unstable conformations as well as by resolving misfolded intermediates. The later observation is related to the function of RNA chaperones. In contrast to RNA chaperones, DEAD-box proteins, however, require an external energy source to release their target RNA molecule after unwinding and many of the RNA helicases depend on a loading platform.Citation63-Citation69 So far, all proteins with RNA chaperone activity described to date fulfill a variety of primary functions in cells,Citation15,Citation29,Citation120,Citation143 but a protein that merely functions as an RNA chaperone has not been identified. Perhaps RNA chaperones were lost during evolution due to the rise of RNA helicases, which proved to be excellent enzymes in numerous cellular processes. Their diversity in reactions and task-dependent regulation by trans-acting ligands, such as other proteins in large RNP machineries, like the spliceosome or during ribosome biogenesis, might have been a selective advantage. Thus, proteins with RNA chaperone activity may be remnants of an ancient world. The finding that a protein known for unwinding of RNA is also able to stabilize RNA structure broadens the spectrum of how DEAD-box proteins are able to shape RNA folding landscapes and to influence RNP architecture and rearrangements. This is of interest specifically in light of the potential function of helicases in nuclear intron splicing, including the spliceosome assembly pathway, and in self-splicing of group II introns, which are considered to be the ancestors of nuclear introns.Citation144
Perspectives
RNA metabolism in yeast mitochondria is tightly associated with the DEAD-box protein Mss116p. This helicase stimulates splicing of structurally distinct introns and is involved in mitochondrial RNA processing and translation.Citation30 Therefore, it is essential that the role of Mss116p in these possibly intertwined processes is defined as much as to explore the mechanism underlying Mss116p function in splicing of the different introns. For example, it has to be addressed whether Mss116p applies distinct mechanisms of action to promote splicing of group I and group II introns, or even for each intron. Thus, it is of utmost importance to dissect the functional relevance of motifs associated with ATP binding and hydrolysis, unwinding and annealing as well as RNA binding in these distinct processes and for the different intron RNAs. Also, Mss116p most likely needs to co-opt with other splicing factors known to assist folding of specific introns.Citation23,Citation27 Given the multiple tasks of Mss116p in yeast mitochondria, deciphering the functional timing is a major milestone along the way of understanding DEAD-box protein-assisted RNA folding and processing as well as translation. Processing and splicing of transcripts intervened by several group I and group II introns is likely to require temporal co-ordination. In other words, folding of an intron might depend on different helicase motifs (and flanking extensions) than refolding of exons to allow splicing of the neighboring intron.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Olga Fedorova and the Waldsich lab for comments on the manuscript. C.W. acknowledges the financial support from the Austrian Science Foundation FWF (grants P23497 and Y401).
References
- Chan RT, Robart AR, Rajashankar KR, Pyle AM, Toor N. Crystal structure of a group II intron in the pre-catalytic state. Nat Struct Mol Biol 2012; 19:555 - 7; http://dx.doi.org/10.1038/nsmb.2270; PMID: 22484319
- Costa M, Michel F. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J 1995; 14:1276 - 85; PMID: 7720718
- Costa M, Déme E, Jacquier A, Michel F. Multiple tertiary interactions involving domain II of group II self-splicing introns. J Mol Biol 1997; 267:520 - 36; http://dx.doi.org/10.1006/jmbi.1996.0882; PMID: 9126835
- Boudvillain M, Pyle AM. Defining functional groups, core structural features and inter-domain tertiary contacts essential for group II intron self-splicing: a NAIM analysis. EMBO J 1998; 17:7091 - 104; http://dx.doi.org/10.1093/emboj/17.23.7091; PMID: 9843513
- Boudvillain M, Delencastre A, Pyle AM. A new RNA tertiary interaction that links active-site domains of a group II intron and anchors them at the site of catalysis. Nature 2000; 406:315 - 8; http://dx.doi.org/10.1038/35018589; PMID: 10917534
- Fedorova O, Pyle AM. Linking the group II intron catalytic domains: tertiary contacts and structural features of domain 3. EMBO J 2005; 24:3906 - 16; http://dx.doi.org/10.1038/sj.emboj.7600852; PMID: 16252007
- Harris-Kerr CL, Zhang M, Peebles CL. The phylogenetically predicted base-pairing interaction between α and α’ is required for group II splicing in vitro.. Proc Natl Acad Sci U S A 1993; 90:10658 - 62; http://dx.doi.org/10.1073/pnas.90.22.10658; PMID: 7504276
- Michel F, Ferat J-L. Structure and activities of group II introns. Annu Rev Biochem 1995; 64:435 - 61; http://dx.doi.org/10.1146/annurev.bi.64.070195.002251; PMID: 7574489
- Toor N, Keating KS, Fedorova O, Rajashankar K, Wang J, Pyle AM. Tertiary architecture of the Oceanobacillus iheyensis group II intron. RNA 2010; 16:57 - 69; http://dx.doi.org/10.1261/rna.1844010; PMID: 19952115
- Toor N, Keating KS, Pyle AM. Structural insights into RNA splicing. Curr Opin Struct Biol 2009; 19:260 - 6; http://dx.doi.org/10.1016/j.sbi.2009.04.002; PMID: 19443210
- Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science 2008; 320:77 - 82; http://dx.doi.org/10.1126/science.1153803; PMID: 18388288
- Toor N, Rajashankar K, Keating KS, Pyle AM. Structural basis for exon recognition by a group II intron. Nat Struct Mol Biol 2008; 15:1221 - 2; http://dx.doi.org/10.1038/nsmb.1509; PMID: 18953333
- Baird NJ, Fang XW, Srividya N, Pan T, Sosnick TR. Folding of a universal ribozyme: the ribonuclease P RNA. Q Rev Biophys 2007; 40:113 - 61; http://dx.doi.org/10.1017/S0033583507004623; PMID: 17931443
- Pyle AM, Fedorova O, Waldsich C. Folding of group II introns: a model system for large, multidomain RNAs?. Trends Biochem Sci 2007; 32:138 - 45; http://dx.doi.org/10.1016/j.tibs.2007.01.005; PMID: 17289393
- Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat Rev Mol Cell Biol 2004; 5:908 - 19; http://dx.doi.org/10.1038/nrm1497; PMID: 15520810
- Shcherbakova I, Mitra S, Laederach A, Brenowitz M. Energy barriers, pathways, and dynamics during folding of large, multidomain RNAs. Curr Opin Chem Biol 2008; 12:655 - 66; http://dx.doi.org/10.1016/j.cbpa.2008.09.017; PMID: 18926923
- Sosnick TR, Pan T. RNA folding: models and perspectives. Curr Opin Struct Biol 2003; 13:309 - 16; http://dx.doi.org/10.1016/S0959-440X(03)00066-6; PMID: 12831881
- Treiber DK, Williamson JR. Exposing the kinetic traps in RNA folding. Curr Opin Struct Biol 1999; 9:339 - 45; http://dx.doi.org/10.1016/S0959-440X(99)80045-1; PMID: 10361090
- Treiber DK, Williamson JR. Beyond kinetic traps in RNA folding. Curr Opin Struct Biol 2001; 11:309 - 14; http://dx.doi.org/10.1016/S0959-440X(00)00206-2; PMID: 11406379
- Woodson SA. Recent insights on RNA folding mechanisms from catalytic RNA. Cell Mol Life Sci 2000; 57:796 - 808; http://dx.doi.org/10.1007/s000180050042; PMID: 10892344
- Woodson SA. Structure and assembly of group I introns. Curr Opin Struct Biol 2005; 15:324 - 30; http://dx.doi.org/10.1016/j.sbi.2005.05.007; PMID: 15922592
- Woodson SA. Compact intermediates in RNA folding. Annu Rev Biophys 2010; 39:61 - 77; http://dx.doi.org/10.1146/annurev.biophys.093008.131334; PMID: 20192764
- Lambowitz AM, Caprara MG, Zimmerly S, Perlman PS. Group I and group II ribozymes as RNPs: Clues to the past and guides to the future. In: Gestland RF, Atkins JF, eds. The RNA world. Cold Spring Harbor: Cold Spring Harbor Laboratory Press, 1999:451-485.
- Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet 2004; 38:1 - 35; http://dx.doi.org/10.1146/annurev.genet.38.072902.091600; PMID: 15568970
- Lehmann K, Schmidt U. Group II introns: structure and catalytic versatility of large natural ribozymes. Crit Rev Biochem Mol Biol 2003; 38:249 - 303; http://dx.doi.org/10.1080/713609236; PMID: 12870716
- Pyle AM, Lambowitz AM. Group II introns: ribozymes that splice RNA and invade DNA. In: Gesteland RF, Cech TR, Atkins JF, eds. The RNA World. Cold Spring Harbor, New York: Cold Spring Harbor Laborartory Press, 2006:469-534.
- Solem A, Zingler N, Pyle AM. J. L-P-T. Group II introns and their protein collaborators. In: Walter NG, Woodson SA, Batey RT, eds. Non-protein coding RNAs. Berlin Heidelberg: Springer-Verlag, 2009:167-182.
- Weeks KM. Protein-facilitated RNA folding. Curr Opin Struct Biol 1997; 7:336 - 42; http://dx.doi.org/10.1016/S0959-440X(97)80048-6; PMID: 9204274
- Zemora G, Waldsich C. RNA folding in living cells. RNA Biol 2010; 7:634 - 41; http://dx.doi.org/10.4161/rna.7.6.13554; PMID: 21045541
- Huang HR, Rowe CE, Mohr S, Jiang Y, Lambowitz AM, Perlman PS. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc Natl Acad Sci U S A 2005; 102:163 - 8; http://dx.doi.org/10.1073/pnas.0407896101; PMID: 15618406
- Séraphin B, Boulet A, Simon M, Faye G. Construction of a yeast strain devoid of mitochondrial introns and its use to screen nuclear genes involved in mitochondrial splicing. Proc Natl Acad Sci U S A 1987; 84:6810 - 4; http://dx.doi.org/10.1073/pnas.84.19.6810; PMID: 3309947
- Séraphin B, Simon M, Boulet A, Faye G. Mitochondrial splicing requires a protein from a novel helicase family. Nature 1989; 337:84 - 7; http://dx.doi.org/10.1038/337084a0; PMID: 2535893
- Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem 1995; 270:20871 - 4; PMID: 7545662
- Adilakshmi T, Bellur DL, Woodson SA. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature 2008; 455:1268 - 72; http://dx.doi.org/10.1038/nature07298; PMID: 18784650
- Bassi GS, de Oliveira DM, White MF, Weeks KM. Recruitment of intron-encoded and co-opted proteins in splicing of the bI3 group I intron RNA. Proc Natl Acad Sci U S A 2002; 99:128 - 33; http://dx.doi.org/10.1073/pnas.012579299; PMID: 11773622
- Caprara MG, Lehnert V, Lambowitz AM, Westhof E. A tyrosyl-tRNA synthetase recognizes a conserved tRNA-like structural motif in the group I intron catalytic core. Cell 1996; 87:1135 - 45; http://dx.doi.org/10.1016/S0092-8674(00)81807-3; PMID: 8978617
- Dai L, Chai D, Gu SQ, Gabel J, Noskov SY, Blocker FJ, et al. A three-dimensional model of a group II intron RNA and its interaction with the intron-encoded reverse transcriptase. Mol Cell 2008; 30:472 - 85; http://dx.doi.org/10.1016/j.molcel.2008.04.001; PMID: 18424209
- Matsuura M, Saldanha R, Ma H, Wank H, Yang J, Mohr G, et al. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev 1997; 11:2910 - 24; http://dx.doi.org/10.1101/gad.11.21.2910; PMID: 9353259
- Paukstelis PJ, Chen JH, Chase E, Lambowitz AM, Golden BL. Structure of a tyrosyl-tRNA synthetase splicing factor bound to a group I intron RNA. Nature 2008; 451:94 - 7; http://dx.doi.org/10.1038/nature06413; PMID: 18172503
- Paukstelis PJ, Coon R, Madabusi L, Nowakowski J, Monzingo A, Robertus J, et al. A tyrosyl-tRNA synthetase adapted to function in group I intron splicing by acquiring a new RNA binding surface. Mol Cell 2005; 17:417 - 28; http://dx.doi.org/10.1016/j.molcel.2004.12.026; PMID: 15694342
- Talkington MW, Siuzdak G, Williamson JR. An assembly landscape for the 30S ribosomal subunit. Nature 2005; 438:628 - 32; http://dx.doi.org/10.1038/nature04261; PMID: 16319883
- Waldsich C, Grossberger R, Schroeder R. RNA chaperone StpA loosens interactions of the tertiary structure in the td group I intron in vivo.. Genes Dev 2002; 16:2300 - 12; http://dx.doi.org/10.1101/gad.231302; PMID: 12208852
- Waldsich C, Masquida B, Westhof E, Schroeder R. Monitoring intermediate folding states of the td group I intron in vivo.. EMBO J 2002; 21:5281 - 91; http://dx.doi.org/10.1093/emboj/cdf504; PMID: 12356744
- Webb AE, Weeks KM. A collapsed state functions to self-chaperone RNA folding into a native ribonucleoprotein complex. Nat Struct Biol 2001; 8:135 - 40; http://dx.doi.org/10.1038/84124; PMID: 11175902
- Weeks KM, Cech TR. Assembly of a ribonucleoprotein catalyst by tertiary structure capture. Science 1996; 271:345 - 8; http://dx.doi.org/10.1126/science.271.5247.345; PMID: 8553068
- Pan C, Russell R. Roles of DEAD-box proteins in RNA and RNP Folding. RNA Biol 2010; 7:667 - 76; http://dx.doi.org/10.4161/rna.7.6.13571; PMID: 21045543
- Bhaskaran H, Russell R. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature 2007; 449:1014 - 8; http://dx.doi.org/10.1038/nature06235; PMID: 17960235
- Bifano AL, Caprara MGA. A DExH/D-box protein coordinates the two steps of splicing in a group I intron. J Mol Biol 2008; 383:667 - 82; http://dx.doi.org/10.1016/j.jmb.2008.08.070; PMID: 18789947
- Bifano AL, Turk EM, Caprara MG. Structure-guided mutational analysis of a yeast DEAD-box protein involved in mitochondrial RNA splicing. J Mol Biol 2010; 398:429 - 43; http://dx.doi.org/10.1016/j.jmb.2010.03.025; PMID: 20307546
- Del Campo M, Mohr S, Jiang Y, Jia H, Jankowsky E, Lambowitz AM. Unwinding by local strand separation is critical for the function of DEAD-box proteins as RNA chaperones. J Mol Biol 2009; 389:674 - 93; http://dx.doi.org/10.1016/j.jmb.2009.04.043; PMID: 19393667
- Del Campo M, Tijerina P, Bhaskaran H, Mohr S, Yang Q, Jankowsky E, et al. Do DEAD-box proteins promote group II intron splicing without unwinding RNA?. Mol Cell 2007; 28:159 - 66; http://dx.doi.org/10.1016/j.molcel.2007.07.028; PMID: 17936712
- Fedorova O, Pyle AM. The brace for a growing scaffold: mss116 protein promotes RNA folding by stabilizing an early assembly intermediate. J Mol Biol 2012; 422:347 - 65; http://dx.doi.org/10.1016/j.jmb.2012.05.037; PMID: 22705286
- Fedorova O, Solem A, Pyle AM. Protein-facilitated folding of group II intron ribozymes. J Mol Biol 2010; 397:799 - 813; http://dx.doi.org/10.1016/j.jmb.2010.02.001; PMID: 20138894
- Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz AM. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J Mol Biol 2007; 365:835 - 55; http://dx.doi.org/10.1016/j.jmb.2006.09.083; PMID: 17081564
- Karunatilaka KS, Solem A, Pyle AM, Rueda D. Single-molecule analysis of Mss116-mediated group II intron folding. Nature 2010; 467:935 - 9; http://dx.doi.org/10.1038/nature09422; PMID: 20944626
- Liebeg A, Mayer O, Waldsich C. DEAD-box protein facilitated RNA folding in vivo.. RNA Biol 2010; 7:803 - 11; http://dx.doi.org/10.4161/rna.7.6.13484; PMID: 21045551
- Mohr S, Matsuura M, Perlman PS, Lambowitz AM. A DEAD-box protein alone promotes group II intron splicing and reverse splicing by acting as an RNA chaperone. Proc Natl Acad Sci U S A 2006; 103:3569 - 74; http://dx.doi.org/10.1073/pnas.0600332103; PMID: 16505350
- Potratz JP, Del Campo M, Wolf RZ, Lambowitz AM, Russell R. ATP-dependent roles of the DEAD-box protein Mss116p in group II intron splicing in vitro and in vivo.. J Mol Biol 2011; 411:661 - 79; http://dx.doi.org/10.1016/j.jmb.2011.05.047; PMID: 21679717
- Sinan S, Yuan X, Russell R. The Azoarcus group I intron ribozyme misfolds and is accelerated for refolding by ATP-dependent RNA chaperone proteins. J Biol Chem 2011; 286:37304 - 12; http://dx.doi.org/10.1074/jbc.M111.287706; PMID: 21878649
- Solem A, Zingler N, Pyle AM. A DEAD protein that activates intron self-splicing without unwinding RNA. Mol Cell 2006; 24:611 - 7; http://dx.doi.org/10.1016/j.molcel.2006.10.032; PMID: 17188036
- Tijerina P, Bhaskaran H, Russell R. Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc Natl Acad Sci U S A 2006; 103:16698 - 703; http://dx.doi.org/10.1073/pnas.0603127103; PMID: 17075070
- Zingler N, Solem A, Pyle AM. Dual roles for the Mss116 cofactor during splicing of the ai5γ group II intron. Nucleic Acids Res 2010; 38:6602 - 9; http://dx.doi.org/10.1093/nar/gkq530; PMID: 20554854
- Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene 2006; 367:17 - 37; http://dx.doi.org/10.1016/j.gene.2005.10.019; PMID: 16337753
- Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol 2010; 20:313 - 24; http://dx.doi.org/10.1016/j.sbi.2010.03.011; PMID: 20456941
- Hilbert M, Karow AR, Klostermeier D. The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol Chem 2009; 390:1237 - 50; http://dx.doi.org/10.1515/BC.2009.135; PMID: 19747077
- Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem Sci 2011; 36:19 - 29; http://dx.doi.org/10.1016/j.tibs.2010.07.008; PMID: 20813532
- Jankowsky E, Fairman ME. RNA helicases--one fold for many functions. Curr Opin Struct Biol 2007; 17:316 - 24; http://dx.doi.org/10.1016/j.sbi.2007.05.007; PMID: 17574830
- Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol 2011; 12:505 - 16; http://dx.doi.org/10.1038/nrm3154; PMID: 21779027
- Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys 2008; 37:317 - 36; http://dx.doi.org/10.1146/annurev.biophys.37.032807.125908; PMID: 18573084
- Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry 2005; 44:13591 - 601; http://dx.doi.org/10.1021/bi0508946; PMID: 16216083
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem 2007; 76:23 - 50; http://dx.doi.org/10.1146/annurev.biochem.76.052305.115300; PMID: 17506634
- Del Campo M, Lambowitz AM. Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol Cell 2009; 35:598 - 609; http://dx.doi.org/10.1016/j.molcel.2009.07.032; PMID: 19748356
- Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science 2006; 313:1968 - 72; http://dx.doi.org/10.1126/science.1131981; PMID: 16931718
- Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 2006; 126:713 - 25; http://dx.doi.org/10.1016/j.cell.2006.08.006; PMID: 16923391
- Fan JS, Cheng Z, Zhang J, Noble C, Zhou Z, Song H, et al. Solution and crystal structures of mRNA exporter Dbp5p and its interaction with nucleotides. J Mol Biol 2009; 388:1 - 10; http://dx.doi.org/10.1016/j.jmb.2009.03.004; PMID: 19281819
- Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa.. Cell 2006; 125:287 - 300; http://dx.doi.org/10.1016/j.cell.2006.01.054; PMID: 16630817
- Theissen B, Karow AR, Köhler J, Gubaev A, Klostermeier D. Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc Natl Acad Sci U S A 2008; 105:548 - 53; http://dx.doi.org/10.1073/pnas.0705488105; PMID: 18184816
- Collins R, Karlberg T, Lehtiö L, Schütz P, van den Berg S, Dahlgren LG, et al. The DEXD/H-box RNA helicase DDX19 is regulated by an alpha-helical switch. J Biol Chem 2009; 284:10296 - 300; http://dx.doi.org/10.1074/jbc.C900018200; PMID: 19244245
- von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol 2009; 16:247 - 54; http://dx.doi.org/10.1038/nsmb.1561; PMID: 19219046
- Mohr G, Del Campo M, Turner KG, Gilman B, Wolf RZ, Lambowitz AM. High-throughput genetic identification of functionally important regions of the yeast DEAD-box protein Mss116p. J Mol Biol 2011; 413:952 - 72; http://dx.doi.org/10.1016/j.jmb.2011.09.015; PMID: 21945532
- Mohr G, Del Campo M, Mohr S, Yang Q, Jia H, Jankowsky E, et al. Function of the C-terminal domain of the DEAD-box protein Mss116p analyzed in vivo and in vitro.. J Mol Biol 2008; 375:1344 - 64; http://dx.doi.org/10.1016/j.jmb.2007.11.041; PMID: 18096186
- Banroques J, Doère M, Dreyfus M, Linder P, Tanner NK. Motif III in superfamily 2 “helicases” helps convert the binding energy of ATP into a high-affinity RNA binding site in the yeast DEAD-box protein Ded1. J Mol Biol 2010; 396:949 - 66; http://dx.doi.org/10.1016/j.jmb.2009.12.025; PMID: 20026132
- Karow AR, Klostermeier D. A conformational change in the helicase core is necessary but not sufficient for RNA unwinding by the DEAD box helicase YxiN. Nucleic Acids Res 2009; 37:4464 - 71; http://dx.doi.org/10.1093/nar/gkp397; PMID: 19474341
- Linder P. Dead-box proteins: a family affair--active and passive players in RNP-remodeling. Nucleic Acids Res 2006; 34:4168 - 80; http://dx.doi.org/10.1093/nar/gkl468; PMID: 16936318
- Karow AR, Klostermeier D. A structural model for the DEAD box helicase YxiN in solution: localization of the RNA binding domain. J Mol Biol 2010; 402:629 - 37; http://dx.doi.org/10.1016/j.jmb.2010.07.049; PMID: 20691700
- Schütz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, et al. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci U S A 2008; 105:9564 - 9; http://dx.doi.org/10.1073/pnas.0800418105; PMID: 18606994
- Klostermeier D, Rudolph MG. A novel dimerization motif in the C-terminal domain of the Thermus thermophilus DEAD box helicase Hera confers substantial flexibility. Nucleic Acids Res 2009; 37:421 - 30; http://dx.doi.org/10.1093/nar/gkn947; PMID: 19050012
- Rudolph MG, Klostermeier D. The Thermus thermophilus DEAD box helicase Hera contains a modified RNA recognition motif domain loosely connected to the helicase core. RNA 2009; 15:1993 - 2001; http://dx.doi.org/10.1261/rna.1820009; PMID: 19710183
- Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell 2007; 28:253 - 63; http://dx.doi.org/10.1016/j.molcel.2007.08.016; PMID: 17964264
- Bizebard T, Ferlenghi I, Iost I, Dreyfus M. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry 2004; 43:7857 - 66; http://dx.doi.org/10.1021/bi049852s; PMID: 15196029
- Rogers GW Jr., Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem 1999; 274:12236 - 44; http://dx.doi.org/10.1074/jbc.274.18.12236; PMID: 10212190
- Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol 2006; 13:981 - 6; http://dx.doi.org/10.1038/nsmb1165; PMID: 17072313
- Rogers GW Jr., Lima WF, Merrick WC. Further characterization of the helicase activity of eIF4A. Substrate specificity. J Biol Chem 2001; 276:12598 - 608; http://dx.doi.org/10.1074/jbc.M007560200; PMID: 11278350
- Pyle AM. RNA helicases and remodeling proteins. Curr Opin Chem Biol 2011; 15:636 - 42; http://dx.doi.org/10.1016/j.cbpa.2011.07.019; PMID: 21862383
- Chen Y, Potratz JP, Tijerina P, Del Campo M, Lambowitz AM, Russell R. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc Natl Acad Sci U S A 2008; 105:20203 - 8; http://dx.doi.org/10.1073/pnas.0811075106; PMID: 19088196
- Henn A, Bradley MJ, De La Cruz EM. ATP utilization and RNA conformational rearrangement by DEAD-box proteins. Annu Rev Biophys 2012; 41:247 - 67; http://dx.doi.org/10.1146/annurev-biophys-050511-102243; PMID: 22404686
- Henn A, Cao W, Licciardello N, Heitkamp SE, Hackney DD, De La Cruz EM. Pathway of ATP utilization and duplex rRNA unwinding by the DEAD-box helicase, DbpA. Proc Natl Acad Sci U S A 2010; 107:4046 - 50; http://dx.doi.org/10.1073/pnas.0913081107; PMID: 20160110
- Aregger R, Klostermeier D. The DEAD box helicase YxiN maintains a closed conformation during ATP hydrolysis. Biochemistry 2009; 48:10679 - 81; http://dx.doi.org/10.1021/bi901278p; PMID: 19839642
- Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci U S A 2008; 105:20209 - 14; http://dx.doi.org/10.1073/pnas.0811115106; PMID: 19088201
- Henn A, Cao W, Hackney DD, De La Cruz EM. The ATPase cycle mechanism of the DEAD-box rRNA helicase, DbpA. J Mol Biol 2008; 377:193 - 205; http://dx.doi.org/10.1016/j.jmb.2007.12.046; PMID: 18237742
- Cao W, Coman MM, Ding S, Henn A, Middleton ER, Bradley MJ, et al. Mechanism of Mss116 ATPase reveals functional diversity of DEAD-Box proteins. J Mol Biol 2011; 409:399 - 414; http://dx.doi.org/10.1016/j.jmb.2011.04.004; PMID: 21501623
- Iost I, Dreyfus M, Linder P. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J Biol Chem 1999; 274:17677 - 83; http://dx.doi.org/10.1074/jbc.274.25.17677; PMID: 10364207
- Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, et al. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science 2004; 304:730 - 4; http://dx.doi.org/10.1126/science.1095596; PMID: 15118161
- Jankowsky E, Bowers H. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res 2006; 34:4181 - 8; http://dx.doi.org/10.1093/nar/gkl410; PMID: 16935886
- Yang Q, Fairman ME, Jankowsky E. DEAD-box-protein-assisted RNA structure conversion towards and against thermodynamic equilibrium values. J Mol Biol 2007; 368:1087 - 100; http://dx.doi.org/10.1016/j.jmb.2007.02.071; PMID: 17391697
- Valdez BC. Structural domains involved in the RNA folding activity of RNA helicase II/Gu protein. Eur J Biochem 2000; 267:6395 - 402; http://dx.doi.org/10.1046/j.1432-1327.2000.01727.x; PMID: 11029582
- Polach KJ, Uhlenbeck OC. Cooperative binding of ATP and RNA substrates to the DEAD/H protein DbpA. Biochemistry 2002; 41:3693 - 702; http://dx.doi.org/10.1021/bi012062n; PMID: 11888286
- Kos M, Tollervey D. The putative RNA helicase Dbp4p is required for release of the U14 snoRNA from preribosomes in Saccharomyces cerevisiae.. Mol Cell 2005; 20:53 - 64; http://dx.doi.org/10.1016/j.molcel.2005.08.022; PMID: 16209945
- Rocak S, Emery B, Tanner NK, Linder P. Characterization of the ATPase and unwinding activities of the yeast DEAD-box protein Has1p and the analysis of the roles of the conserved motifs. Nucleic Acids Res 2005; 33:999 - 1009; http://dx.doi.org/10.1093/nar/gki244; PMID: 15718299
- Mallam AL, Jarmoskaite I, Tijerina P, Del Campo M, Seifert S, Guo L, et al. Solution structures of DEAD-box RNA chaperones reveal conformational changes and nucleic acid tethering by a basic tail. Proc Natl Acad Sci U S A 2011; 108:12254 - 9; http://dx.doi.org/10.1073/pnas.1109566108; PMID: 21746911
- Fedorova O, Waldsich C, Pyle AM. Group II intron folding under near-physiological conditions: collapsing to the near-native state. J Mol Biol 2007; 366:1099 - 114; http://dx.doi.org/10.1016/j.jmb.2006.12.003; PMID: 17196976
- Su LJ, Waldsich C, Pyle AM. An obligate intermediate along the slow folding pathway of a group II intron ribozyme. Nucleic Acids Res 2005; 33:6674 - 87; http://dx.doi.org/10.1093/nar/gki973; PMID: 16314300
- Waldsich C, Pyle AM. A folding control element for tertiary collapse of a group II intron ribozyme. Nat Struct Mol Biol 2007; 14:37 - 44; http://dx.doi.org/10.1038/nsmb1181; PMID: 17143279
- Waldsich C, Pyle AM. A kinetic intermediate that regulates proper folding of a group II intron RNA. J Mol Biol 2008; 375:572 - 80; http://dx.doi.org/10.1016/j.jmb.2007.10.052; PMID: 18022197
- Liebeg A, Waldsich C. Probing RNA structure within living cells. Methods Enzymol 2009; 468:219 - 38; http://dx.doi.org/10.1016/S0076-6879(09)68011-3; PMID: 20946772
- Cao Y, Woodson SA. Destabilizing effect of an rRNA stem-loop on an attenuator hairpin in the 5′ exon of the Tetrahymena pre-rRNA. RNA 1998; 4:901 - 14; http://dx.doi.org/10.1017/S1355838298980621; PMID: 9701282
- Cao Y, Woodson SA. Refolding of rRNA exons enhances dissociation of the Tetrahymena intron. RNA 2000; 6:1248 - 56; http://dx.doi.org/10.1017/S1355838200000893; PMID: 10999602
- Fang XW, Thiyagarajan P, Sosnick TR, Pan T. The rate-limiting step in the folding of a large ribozyme without kinetic traps. Proc Natl Acad Sci U S A 2002; 99:8518 - 23; http://dx.doi.org/10.1073/pnas.142288399; PMID: 12084911
- Fang XW, Pan T, Sosnick TR. Mg2+-dependent folding of a large ribozyme without kinetic traps. Nat Struct Biol 1999; 6:1091 - 5; http://dx.doi.org/10.1038/70016; PMID: 10581546
- Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, et al. RNA chaperones, RNA annealers and RNA helicases. RNA Biol 2007; 4:118 - 30; http://dx.doi.org/10.4161/rna.4.3.5445; PMID: 18347437
- Nikolcheva T, Woodson SA. Facilitation of group I splicing in vivo: misfolding of the Tetrahymena IVS and the role of ribosomal RNA exons. J Mol Biol 1999; 292:557 - 67; http://dx.doi.org/10.1006/jmbi.1999.3083; PMID: 10497021
- Pan T, Sosnick TR. Intermediates and kinetic traps in the folding of a large ribozyme revealed by circular dichroism and UV absorbance spectroscopies and catalytic activity. Nat Struct Biol 1997; 4:931 - 8; http://dx.doi.org/10.1038/nsb1197-931; PMID: 9360610
- Pan J, Woodson SA. Folding intermediates of a self-splicing RNA: mispairing of the catalytic core. J Mol Biol 1998; 280:597 - 609; http://dx.doi.org/10.1006/jmbi.1998.1901; PMID: 9677291
- Rangan P, Masquida B, Westhof E, Woodson SA. Architecture and folding mechanism of the Azoarcus group I pre-tRNA. J Mol Biol 2004; 339:41 - 51; http://dx.doi.org/10.1016/j.jmb.2004.03.059; PMID: 15123419
- Semrad K, Schroeder R. A ribosomal function is necessary for efficient splicing of the T4 phage thymidylate synthase intron in vivo.. Genes Dev 1998; 12:1327 - 37; http://dx.doi.org/10.1101/gad.12.9.1327; PMID: 9573049
- Bassi GS, Weeks KM. Kinetic and thermodynamic framework for assembly of the six-component bI3 group I intron ribonucleoprotein catalyst. Biochemistry 2003; 42:9980 - 8; http://dx.doi.org/10.1021/bi0346906; PMID: 12924947
- Herbert CJ, Labouesse M, Dujardin G, Slonimski PP. The NAM2 proteins from S. cerevisiae and S. douglasii are mitochondrial leucyl-tRNA synthetases, and are involved in mRNA splicing. EMBO J 1988; 7:473 - 83; PMID: 3284745
- Kaspar BJ, Bifano AL, Caprara MG. A shared RNA-binding site in the Pet54 protein is required for translational activation and group I intron splicing in yeast mitochondria. Nucleic Acids Res 2008; 36:2958 - 68; http://dx.doi.org/10.1093/nar/gkn045; PMID: 18388132
- McGraw P, Tzagoloff A. Assembly of the mitochondrial membrane system. Characterization of a yeast nuclear gene involved in the processing of the cytochrome b pre-mRNA. J Biol Chem 1983; 258:9459 - 68; PMID: 6348045
- Rho SB, Martinis SA. The bI4 group I intron binds directly to both its protein splicing partners, a tRNA synthetase and maturase, to facilitate RNA splicing activity. RNA 2000; 6:1882 - 94; http://dx.doi.org/10.1017/S1355838200001254; PMID: 11142386
- Weeks KM, Cech TR. Protein facilitation of group I intron splicing by assembly of the catalytic core and the 5′ splice site domain. Cell 1995; 82:221 - 30; http://dx.doi.org/10.1016/0092-8674(95)90309-7; PMID: 7628013
- Gampel A, Cech TR. Binding of the CBP2 protein to a yeast mitochondrial group I intron requires the catalytic core of the RNA. Genes Dev 1991; 5:1870 - 80; http://dx.doi.org/10.1101/gad.5.10.1870; PMID: 1916266
- Gampel A, Nishikimi M, Tzagoloff A. CBP2 protein promotes in vitro excision of a yeast mitochondrial group I intron. Mol Cell Biol 1989; 9:5424 - 33; PMID: 2685564
- Gampel A, Tzagoloff A. In vitro splicing of the terminal intervening sequence of Saccharomyces cerevisiae cytochrome b pre-mRNA. Mol Cell Biol 1987; 7:2545 - 51; PMID: 3302680
- Weeks KM, Cech TR. Efficient protein-facilitated splicing of the yeast mitochondrial bI5 intron. Biochemistry 1995; 34:7728 - 38; http://dx.doi.org/10.1021/bi00023a020; PMID: 7540041
- Garcia I, Weeks KM. Structural basis for the self-chaperoning function of an RNA collapsed state. Biochemistry 2004; 43:15179 - 86; http://dx.doi.org/10.1021/bi048626f; PMID: 15568809
- Watts T, Khalimonchuk O, Wolf RZ, Turk EM, Mohr G, Winge DR. Mne1 is a novel component of the mitochondrial splicing apparatus responsible for processing of a COX1 group I intron in yeast. J Biol Chem 2011; 286:10137 - 46; http://dx.doi.org/10.1074/jbc.M110.205625; PMID: 21257754
- Turk EM, Caprara MG. Splicing of yeast aI5beta group I intron requires SUV3 to recycle MRS1 via mitochondrial degradosome-promoted decay of excised intron ribonucleoprotein (RNP). J Biol Chem 2010; 285:8585 - 94; http://dx.doi.org/10.1074/jbc.M109.090761; PMID: 20064926
- Grohman JK, Del Campo M, Bhaskaran H, Tijerina P, Lambowitz AM, Russell R. Probing the mechanisms of DEAD-box proteins as general RNA chaperones: the C-terminal domain of CYT-19 mediates general recognition of RNA. Biochemistry 2007; 46:3013 - 22; http://dx.doi.org/10.1021/bi0619472; PMID: 17311413
- Hilliker A, Gao Z, Jankowsky E, Parker R. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol Cell 2011; 43:962 - 72; http://dx.doi.org/10.1016/j.molcel.2011.08.008; PMID: 21925384
- Mohr S, Stryker JM, Lambowitz AM. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell 2002; 109:769 - 79; http://dx.doi.org/10.1016/S0092-8674(02)00771-7; PMID: 12086675
- Mohr G, Zhang A, Gianelos JA, Belfort M, Lambowitz AM. The neurospora CYT-18 protein suppresses defects in the phage T4 td intron by stabilizing the catalytically active structure of the intron core. Cell 1992; 69:483 - 94; http://dx.doi.org/10.1016/0092-8674(92)90449-M; PMID: 1533818
- Woodson SA. Taming free energy landscapes with RNA chaperones. RNA Biol 2010; 7:677 - 86; http://dx.doi.org/10.4161/rna.7.6.13615; PMID: 21045544
- Michel F, Costa M, Westhof E. The ribozyme core of group II introns: a structure in want of partners. Trends Biochem Sci 2009; 34:189 - 99; http://dx.doi.org/10.1016/j.tibs.2008.12.007; PMID: 19299141