Abstract
The opioid receptors are among the most highly studied members of the superfamily of G-protein coupled receptors. Morphine and endogenous mu opioid peptides exert their pharmacological actions mainly through the mu opioid receptor (MOR). Expression of opioid receptor proteins is controlled by extensive transcriptional and post-transcriptional processing. Previously, the 5′-untranslated region (UTR) of the mouse MOR was found to be important for post-transcriptional regulation of the MOR gene in neuronal cells. Here, we demonstrate for the first time the role of vimentin as a post-transcriptional repressor in MOR gene regulation. To identify potential regulators of the mouse MOR gene, we performed affinity column chromatography using 5′-UTR-specific RNA oligonucleotides using neuroblastoma NS20Y cells. Chromatography was followed by two-dimensional gel electrophoresis and MALDI-TOF mass spectrometry. We identified an intermediate filament protein, vimentin, which bound specifically to the region between -175 and -150 (175–150) of the MOR 5′-UTR. Binding was confirmed by western blot analysis and RNA supershift assay. Furthermore, a cotransfection study demonstrated that the presence of vimentin resulted in reduced expression of the mouse MOR. Our data suggest that vimentin functions as a repressor of MOR translation, dependent on 175–150 of the MOR 5′-UTR.
Introduction
Opioid analgesics are widely used for the treatment of moderate to severe pain. Their efficacy is the result of their ability to mimic endogenous opioid peptides on the opioid receptors (ORs). ORs are classified into three major types (μ, δ and κ) and have been characterized by molecular cloning and in numerous pharmacological reports.Citation1,Citation2 Several studies have suggested that the mu opioid receptor (MOR) plays a key role in mediating the major clinical effects of morphine, as well as in the development of morphine tolerance and physical dependence with chronic administration.Citation3 In addition, numerous studies have identified multiple cis-acting elements within the promoter and 5′-untranslated region (UTR) of MOR.Citation2-Citation14
Gene expression can be regulated at multiple steps, including transcription, mRNA stability and translation. For gene expression to be established at an appropriate level, some of these regulatory processes must be under rapid and precise control. Because transcriptional regulation is not sufficient to determine the level of gene expression and its duration, post-transcriptional regulation is necessary for finely tuned gene expression.Citation15 While interest in post-transcriptional regulation has increased recently with the explosive discoveries of large numbers of non-coding RNAs such as microRNAs, post-transcriptional processes still depend largely on the functions of RNA-binding proteins (RBPs).Citation16 Efficient analysis of RNA can be performed by hybridization or sequencing-based methods; however, in the cellular environment RNA is associated with RBPs, which together form functional ribonucleoprotein (RNP) complexes. In addition, mRNA localization is a complex process that begins with the recognition of cis-acting elements in nascent transcripts by trans-acting RBPs. After the RNP complex is established, it exits the nucleus and is joined by additional cytoplasmic factors to create a RNP granule. The RNP granule associates either with cytoskeleton or with specific membranes, mitochondria and peroxiosomes to facilitate mRNA delivery and possibly, localized translation. Therefore, determination of RBP specificities is a critical step in the elucidation and analysis of mechanisms involved in co- and post-transcriptional gene regulation.Citation17
Vimentin, one of the type III intermediate filament proteins, is a multifunctional cytoskeletal protein. Its ability to interact with a large number of proteins makes it a potential regulator of several physiological functions. However, the true function of vimentin, apart from maintaining the structural integrity of cells, is yet to be determined.Citation18 In this report, we demonstrate that vimentin interacts with the mouse MOR 5′-UTR, specifically with the region between -175 and -150 of the MOR 5′-UTR, which we have named 175–150 for the purposes of this study. We used affinity column chromatography containing a specific competitor, two-dimensional gel electrophoresis, and mass spectrometry to purify and identify factors that interact preferentially with 175–150 RNA from mouse neuronal cells. Our results showed that the wild-type 175–150 sequences from the mouse MOR 5′-UTR region interact specifically with vimentin to regulate mouse MOR gene expression. Thus, we demonstrate that this important cytoskeletal protein serves as a post-transcriptional regulator of the mouse MOR gene.
Results
Deletion of a cis (trans)-element in the 5′-UTR of MOR results in loss of gene expression
Our previous work has shown that mouse MOR expression is inhibited at the translational level by the 5′-untranslated region (UTR) of MOR.Citation8 However, the post-transcriptional mechanisms responsible for MOR regulation in the 5′-UTR region are not well known, and are the focus of this study (). To identify the cis-element and trans-acting factors required for MOR translation activity, we performed a 5′-deletion analysis.
Figure 1. Translation of the mouse MOR gene is controlled by 5′UTR-deletion analysis. (A) Schematic representation of the mouse MOR 5′-UTR. (B and C) Schematic representation of reporter constructs with wild type and deleted mouse MOR 5′-UTRs. The deleted sequences are described in Materials and Methods. Transient transfection of each deleted construct was performed in NS20Y cells. After transfection, cells were trypsinized and half were used for luciferase and β-galactosidase activity assays, while the other half were used for RNA extraction and transcript quantification. Relative LUC activity and mRNA levels were determined as the ratio of LUC/β-gal and LUC/LacZ as described in Materials and Methods. Error bars indicate the standard errors of triplicate LUC assays.
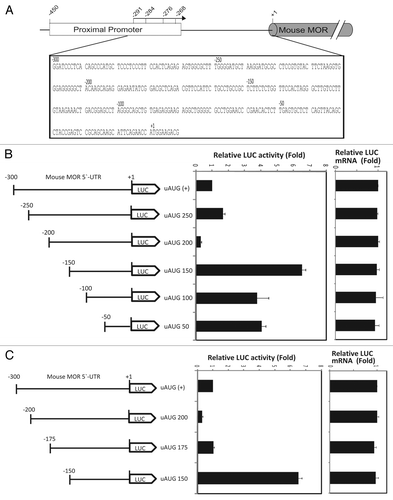
Chimeric constructs ( and C) containing various lengths of the 5′-UTR of mouse MOR were cloned into a pGL3-promoter plasmid containing a luciferase reporter in order to confirm their roles in both transcription and translation by real-time PCR and luciferase assays ( and C). Transcripts levels were very similar among all constructs, indicating that the deletion of nucleotides (nt) did not alter transcription levels. In contrast, LUC activity was differentially affected by the deletion of 5′-UTR sequences. Particularly, the deletion between -200 and -150 nt caused a 6.5-fold increase in LUC activity compared with uAUG (+) expression (see uAUG 200 vs. uAUG 150; ), suggesting the involvement of the -200 to -150 bp region in negative regulation of translational activity. After confirming the role of this 50-bp cis-element, we further narrowed the region of interest to 25 bp (uAUG 175). The deletion between -175 and -150 nt caused the greatest increase in LUC activity (i.e., translational activity but not transcriptional activity) compared with the control (see uAUG 175 vs. uAUG 150; ). Collectively, these results demonstrate that the region between -175 and -150 contains a negative cis-acting and/or trans-acting element binding site that affects MOR gene regulation.
A 54 kDa protein binds to the 5′-UTR of MOR mRNA
In general, many RNA binding proteins (RBPs) remain uncharacterized. Several methods have been developed to identify and characterize RBPs and the RNAs with which they interact. Messenger RNP (mRNP) complexes were initially isolated by several methods.Citation19 Our data show that the 25-bp region between -175 and -150 of the 5′-UTR (175–150) can regulate MOR expression at the post-transcriptional level. We therefore speculated that regulation of MOR expression by 175–150 could involve an interaction between the 5′-UTR of MOR mRNA and certain binding factors. To confirm this inference, we prepared cytosolic extracts from mouse NS20Y cells and tested the ability of protein constituents of these extracts to bind 175–150 mRNA by affinity column assay ().
Figure 2. Schematic representation of the procedure for one-step purification of RNA binding proteins. (A) RNA oligonucleotides containing 25 nucleotide sequences with or without biotin label. (B) Outline of the modified one-step purification of RBPs using an affinity column. RNA oligonucleotides biotinylated on the 5′- or 3′-terminus were used as affinity particles. Cytosolic proteins were added to the affinity particles, incubated and washed. Proteins bound to the particles were released by heating in SDS sample buffer. Competitor experiments to eliminate non-specific proteins and to identify specific binding (filled star) were performed by preincubating the cytosolic proteins with a 2-fold excess of nonbiotinylated RNAs (Non B-175150) as competitors prior to affinity binding. (C) Coomassie-stained gel of RNA binding proteins purified from NS20Y cytosolic extracts with competitor. The competitor RNA sequences are shown in panel A.
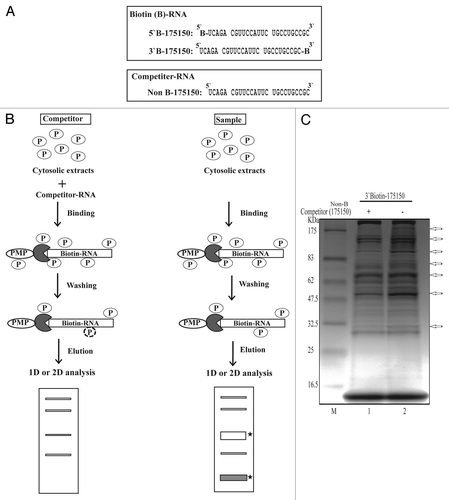
For these studies, we used biotin-labeled RNA oligonucleotides and non-biotinylated competitors () to identify RBPs specific to 175–150. Using this protocol, we were able to purify and identify proteins from NS20Y cytosolic extracts that bound specifically to 175–150 (). After we confirmed 175–150-specific binding by competitor assay, we used 2-dimensional electrophoresis (2-DE) to further characterize the major proteins that bound the 175–150 RNA sequence (). On average, over 50 protein spots were detected at pH 3-pH 10 in the 2-DE images of competitor-treated controls and samples. Most spots were distributed within a pH range of 4–8 with molecular masses of 30–80 kDa. Comparisons of the 2-DE images identified two protein spots present in the sample gels. These spots were analyzed using MALDI-TOF mass spectrometry and bioinformatics. The two spots were identified as vimentin ().
Figure 3. Simply Blue safe-stained 2-DE images of RBPs purified using an affinity column. Purified samples were separated on pH 3–10 IPG strips followed by separation by 4–20% SDS-PAGE. The competitor assay is shown in (A) and the sample assay shown in (B). Molecular weight markers are indicated on the left, and PI values are indicated the bottom. Arrows indicated spots that were subjected to analysis by MALDI-TOF mass spectrometry and bioinformatics. Detailed information for each spot is listed in .
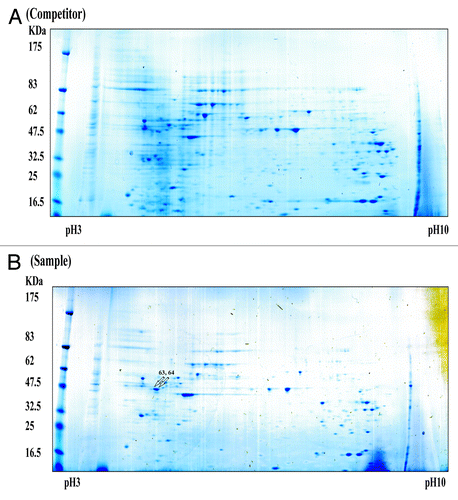
Table 1. Analysis by MALDI-TOF MS of tryptic peptide profiles
Relationship between MOR 5′-UTR and vimentin
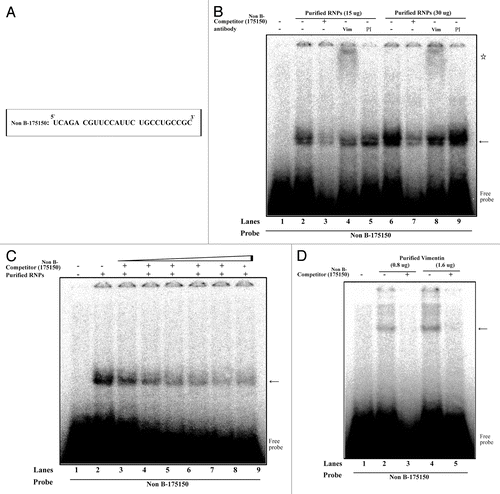
We next used independent methods (western blotting and REMSA) to validate the interaction between vimentin and mouse MOR 175–150. Western blot analyses were performed using purified proteins from NS20Y cells. Our results showed that the purified proteins contained higher amounts of vimentin () compared with the competitive samples, indicating that the competitor (Non-B-175150) decreased the binding activity of vimentin. Furthermore, REMSAs were performed using 32P-labeling methods and are shown in . The REMSA experiments were performed using a 32P-labeled 175–150 RNA probe () and unlabeled purified RNPs by RNA-affinity purification. REMSA results demonstrated that vimentin binds the target 175–150 probe (see lanes 2 and 6 vs. lane 1; ). A 100-fold molar excess of unlabeled 175–150 RNA oligonucleotides (, lanes 3 and 7) inhibited the complex formation. The specificity of the RNA-protein interaction was verified using anti-vimentin antibodies. The vimentin-RNA complexes were supershifted (star shape) by addition of their own antibodies (, lanes 4 and 8), but the pre-immune serum (PI) was not effective in the super-shift assay (, lanes 5 and 9). To confirm the binding of 175–150 RNA probe to vimentin, the purified RNPs were assessed in the presence of varying concentrations (0 ~250 molar excess) of the unlabeled RNA probe () and the purified vimentin was assessed in the presence of the unlabeled probe (). The results showed that an excess of unlabeled RNA probe could displace the radio-labeled probe from the RNA-protein complex. These results suggest the specificity of the binding of the cytoskeletal protein vimentin, to the 175–150 RNA sequences.
Figure 4. Identification of vimentin as a 175–150 RNA binding protein using a competitor via western blot analysis of purified RBPs performed with anti-vimentin antibodies. Vimentin protein levels were measured in NS20Y cells by western blotting after purification. Signal intensities were analyzed using ImageQuant 5.2 software.
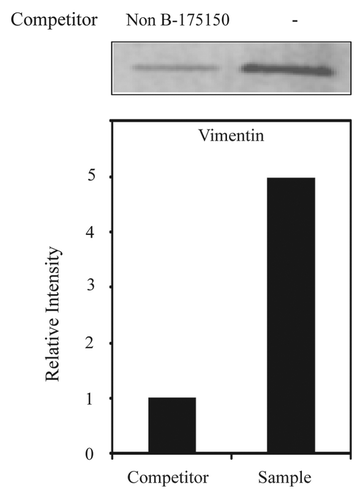
Figure 5. Interaction of vimentin with 175–150 RNA derived from the mouse MOR 5′-UTR. (A) The 25 bp of RNA sequence derived from mouse MOR 5′-UTR (between -175 and -150). (B) REMSAs were performed using 32P-labeled Non B-175150 as a probe with non-labeled purified RNPs by RNA-affinity purification. Lane 1, probe alone; lanes 2 and 5, purified RNPs without antibody; lanes 3 and 7, self-competitor without antibody; lanes 4 and 8, anti-vimentin; lanes 6 and 9, preimmune serum (PI). The arrow indicates the protein-RNA complexes and the star symbol indicates the shifted band corresponding to the probe. (C) Competitive REMSA experiments performed to assess the displacement of the probe complexed with the protein in the presence of excess molar concentrations (10, 50, 100, 150, 200 and 250×) of the unlabelled Non B-175150 oligonucleotide. The arrow indicates the protein-RNA complexes (D) REMSAs were performed using 32P-labeled Non B-175150 as a probe with non-labeled purified vimentin. Lane 1, probe alone; lanes 2 and 4, purified vimentin without competitor; lanes 3 and 5, purified vimentin with self-competitor. The arrow indicates the vimentin-RNA complexes. These data are representative of 2 independent experiments.
Defining the core binding motif
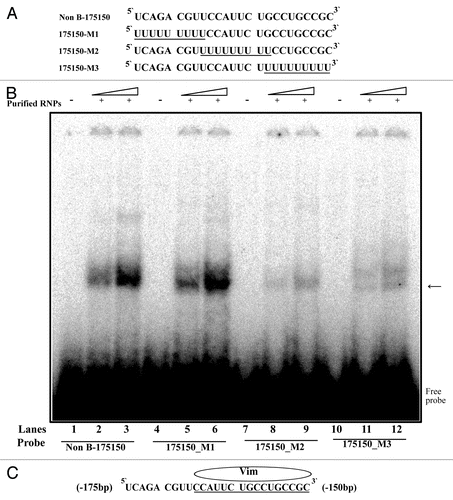
To determine the binding motif within the RNA sequences (175–150) of the MOR 5′-UTR, REMSA were performed using purified RNPs with 32P-labeled wild-type probe (, Non B-175150) and mutated probes as indicated (, 175150-M1 through 175150-M3). The RNA-protein complexes were observed for purified RNPs (vimentin) using Non B-175150 and 175150-M1 (, lanes 2, 3, 5 and 6). No RNA-protein complexes were observed using the 175150-M2 and 175150-M3 sequences (, lanes 8, 9, 11 and 12). This suggests that the RNA sequence 5′-CCAUUCUGCCUGCCGC-3′ (, underlined) served as the vimentin binding motif within the RNA sequence of the mouse MOR 5′-UTR.
Figure 6. REMSA analysis of vimentin binding motif using mutant RNA-oligonucleotide sequences. (A) Representation of RNA-oligonucleotide sequence (Non B-175150) and mutant RNA-oligonucleotide sequences (175150-M1 ~175–150-M3). (B) REMSAs were performed using 32P-labeled wild-type (Non B-175150) and mutated oligonucleotides (175150-M1, M2 and M3) as probes with non-labeled purified RNPs by RNA-affinity purification. Lanes 1, 4, 7 and 10, probe alone; lanes 2, 5, 8 and 11, purified RNPs (15 ug) with indicated probe; lanes 3, 6, 9 and 12, purified RNPs (30 ug) with indicated probe. The arrow indicates the protein-RNA complexes. (C) Schematic representation of the binding motif for vimentin on the RNA sequence. These data are representative of 2 independent experiments.
Vimentin negatively regulates mouse MOR gene expression by 175–150 in the 5′-UTR of MOR
Our western and REMSA data demonstrate that vimentin binds to the wild type 175–150 RNA oligonucleotides derived from the 5′-UTR of mouse MOR. To examine the functional role of vimentin in normal mouse MOR regulation, we used a uAUG (+) reporter construct containing the mouse 5′-UTR region (minus promoter) fused in-frame to a luciferase reporter construct (). Luciferase assays and real-time PCR were performed with vimentin protein and RNAs from NS20Y and Neuro2A cells that had been transfected with varying amounts (0–4 μg) of vimentin protein expression vectors and the normal mouse uAUG (+) reporter construct. As shown in , the protein levels of luciferase were decreased by co-transfection with vimentin compared with uAUG (+) construct alone. Furthermore, vimentin expression downregulated luciferase gene expression at the post-transcriptional level in a dose-dependent manner in both NS20Y and Neuro2A cells.
Figure 7. Mouse MOR expression levels in vimentin-transfected NS20Y and Neuro2A cells. Graphic representation of relative luciferase expression determined by luciferase assay (Promega) of the constructs shown in [uAUG (+)] with 2–4 μg of RBP expression constructs. Relative LUC activity and mRNA levels were determined as the ratio of LUC/β-gal and LUC/LacZ as described in Materials and Methods. *P and #P < 0.05 compared with non-treated control.
![Figure 7. Mouse MOR expression levels in vimentin-transfected NS20Y and Neuro2A cells. Graphic representation of relative luciferase expression determined by luciferase assay (Promega) of the constructs shown in Figure 1 [uAUG (+)] with 2–4 μg of RBP expression constructs. Relative LUC activity and mRNA levels were determined as the ratio of LUC/β-gal and LUC/LacZ as described in Materials and Methods. *P and #P < 0.05 compared with non-treated control.](/cms/asset/f1d8b123-123c-4baf-968a-141574e705f3/krnb_a_10923022_f0007.gif)
Discussion
Protein levels in cells are regulated not only by the rate of transcription but also by rates of subsequent events, including RNA processing, nuclear RNA export, translation and RNA decay.Citation20 These post-transcriptional events are controlled by RNA-binding proteins (RBPs). Many studies have suggested that RBPs indirectly couple transcriptional and subsequent post-transcriptional steps by interacting with their target transcripts.Citation21 Although progressive coupling of these processes is widely accepted, both coupling and coordination are important in determining how, when and where functionally related subpopulations of mRNAs are translated.Citation22 RBPs are universal in living cells, and are estimated to number approximately 650 in yeast and over 2500 in mammals.Citation23,Citation24 Although some RBPs are thought to bind RNA with little or no sequence specificity, many and possibly most RBPs specifically bind to distinct subpopulations of RNA.Citation16 To date, existing RNA tags can be classified into two categories: tags developed based on RNA-protein interactions found in nature, and tags developed from apatamers evolved in vitro to possess affinity for specific ligands such as streptavidin, streptomycin or tobramycin.Citation25
In the present study, we analyzed the binding region of trans (cis)-acting elements in the mouse MOR 5′-UTR by 5′-deletion analysis. We also investigated the interaction of trans (cis)-acting elements within mouse MOR 5′-UTR RNA and their importance in post-transcriptional regulation of MOR. We identified a new candidate RBP (vimentin) associated with 175–150 from the mouse MOR 5′-UTR using streptavidin paramagnetic particles (PMP). We used typical methods (RNA affinity purification assays with or without competition) to isolate mRNA bound to RBPs. Western analyses and REMSAs further revealed the characteristics of the sequence-specific interaction between vimentin and 175–150 of the mouse MOR 5′-UTR. In particular, the 16-base sequence (-166 to -150, 5′-CCAUUCUGCCUGCCGC-3′) is critical for vimentin-RNA complex formation. Functional analyses suggest that vimentin regulates the MOR 5′-UTR containing wild type RNA sequences. These data suggest that vimentin acts as post-transcriptional repressor. These results were confirmed by studies in the mouse neuroblastoma cell lines NS20Y and Neuro2A.
The cytoskeletal system is essential for maintaining various cellular functions. However, the newly elucidated properties of intermediate filaments (IFs) have important functional implications for their involvement in cell division, protein trafficking, cellular motility, intracellular signaling, and the regional control of cytoplasmic architecture.Citation26 In mammalian cells, there are three types of cytoskeletal filaments: actin-containing microfilaments, tubulin-containing microtubules and IFs. Among them, vimentin is a major component of IFs in cells of mesenchymal tissues.Citation27
Our data suggest that vimentin regulates the mouse MOR gene containing wild type 175–150 RNA sequences. These results were confirmed by our studies in NS20Y and Neuro2A cells, mouse neuroblastoma cell lines which endogenously express the MOR gene. Increasing the exogenous expression of vimentin in these cells downregulated MOR expression in a dose-dependent manner. Taken together, these results indicate that vimentin acts as a repressor of mouse MOR expression in neuronal cells via a mechanism dependent on 175–150 of the mouse MOR. Our study offers a new perspective on the role of vimentin in MOR gene regulation and will facilitate a better understanding of the molecular mechanisms underlying MOR expression.
Materials and Methods
Plasmid construction
The luciferase fusion constructs uAUG (+) used here have been described previously.Citation8,Citation12 The divided constructs (uAUG 250, uAUG 200, uAUG 175, uAUG 150, uAUG 100 and uAUG 50) were generated by PCR using the following primers: uAUG 250: 5′-ATAAGCTTTGGGGATGCTAAGGATGCG-3′ (forward); uAUG 200: 5′-CAAAGCTTACAAGCAGAGGAGAATATC-3′ (forward); uAUG 175: 5′-CTAAGCTTTCAGACGTTCCATTCTGCC-3′ (forward); uAUG 150: 5′-CTAAGCTTTCTTCTCTGGTTCCACTAG-3′ (forward); uAUG 100: 5′-TAAAGCTTAGGGCAGCTGTGAGAGGAA-3′ (forward); uAUG 50: 5′-GTAAGCTTTGAGTGCTCTCAGTTACAG-3′ (forward) and GL2: 5′-CTTTATGTTTTTGGCGTCTTCC-3′ (reverse). All DNA inserts were subcloned into the pGL3-promoter vector using HindIII and NcoI sites created with the primers. To clone the vimentin gene, total RNA was isolated from mouse NS20Y cells and treated with RNase-free DNase (Promega). RT-PCR was performed with primers designed using the gene sequence information for the protein vimentin (Gene ID: M24849.1) as follows: 5′-CTAAGCTTAAGCCATGTCTACCAGGT-3′ (forward) and 5′-CTTCTAGATTCAAGGTCATCGTGATG-3′ (reverse). The PCR conditions were as follows: 94°C for 5 min; 35 cycles of 94°C for 1 min, 60°C for 1 min, 72°C for 1.5 min; and 72°C for 10 min. The RT-PCR products were excised from 1% agarose gel purified using a QIAQuick gel extraction kit (Qiagen) and subcloned into the pcDNA-4A vector using HindIII and XbaI sites created with the primers. All constructs were confirmed by DNA sequencing.
Cell culture, DNA transfection and reporter gene assay
Mouse neuroblastoma cell lines NS20Y and Neuro2A were grown in Dulbecco’s minimum essential medium supplemented with 10% heat-inactivated fetal bovine serum at 37°C in a humidified atmosphere of 5% CO2. Transfection and reporter gene assays were performed as described previously.Citation7,Citation8
Quantification of LUC and LacZ transcripts by real-time PCR and reverse transcription (RT)-PCR
The above assays were performed as described previously.Citation8
Cytosolic extract preparation
Extract preparation was performed as described previously.Citation28 Briefly, cells were lysed in cytoplasmic extract buffer (CEB) (10 mM HEPES-KOH, pH 7.6, 100 mM KCl, 2.5 mM MgCl2, 1 mM dithiothreitol [DTT], 0.25% NP-40, EDTA-free Complete Mini protease inhibitor, 1 mM NaF and 1 mM Na3VO4). The lysates were centrifuged at 16,200 × g for 15 min at 4°C and the supernatants were used for RNA-affinity purification.
RNA-affinity purification
RNA-affinity purification was performed as described in . The following procedure is based on the interaction between biotin and streptavidin. RNA oligonucleotides were synthesized and purified using HPLC. In a sterile tube, 500 µl of 0.5X SSC solution was added to 500 pmol of each 3′-terminal-biotinylated RNA. Meanwhile, 500 pmol of streptavidin-paramagnetic particles (Promega) were resuspended by gently flicking the bottom of the tube until they were completely dispersed, then captured by placing the tube in a magnetic stand. The supernatant was carefully removed. The magnetic particles were washed three times with 0.5X SSC and resuspended in 100 µl of 0.5X SSC. Five hundred pmol of biotinylated RNA and 500 pmol of streptavidin-paramagnetic particles were combined and incubated for 15 min at room temperature. Samples were mixed by gentle inversion every 2 min. The magnetic beads were captured using a magnetic stand. The particles were washed three times with 300 µl of CEB buffer 1 (10 mM HEPES-KOH, 2.5 mM MgCl2, 100 mM KCl, 1 mM DTT, 0.25% NP-40, 1 mM NaF, 1 mM Na3VO4 and 1X protease inhibitors), pH 7.6. One mg of cytosolic proteins was added to the affinity particles and incubated for 1 h at 4°C. The particles were washed three times each with CEB buffer 1 and CEB buffer 2 (10 mM HEPES-KOH, 2.5 mM MgCl2, 200 mM KCl, 1 mM DTT, 0.25% NP-40, 1 mM NaF, 1 mM Na3VO4 and 1X protease inhibitors), pH 7.6. Proteins bound to the particles were released by incubation in 50 µl 1X SDS sample buffer for 10 min at 95°C in a heating block.
In order to eliminate cytosolic proteins that might bind non-specifically, control experiments were performed as follows: 1000 pmol of non-biotinylated RNA (2X competitor) were mixed with 1 mg of cytosolic proteins for 15 min on ice. The cytosolic extracts containing the 2X competitor were added to the affinity particles and incubated for 1 h at 4°C. The remainder of the procedure was performed as above. The resultant protein solutions with and without competitor were electrophoresed on a 4–20% gradient gel (Invitrogen) and stained with Coomassie blue (Simply Blue Safe-Stain, Invitrogen).
Immunoblot analysis
Purified proteins were resolved by SDS-PAGE using a 4–20% gradient polyacrylamide gel (Invitrogen). Gels were electroblotted onto polyvinylidene difluoride membranes (Amersham Bioscience) in transfer buffer (48 mM TRIS-HCl, 39 mM glycine and 20% methanol). Membranes were blocked in a solution of 5% dry milk and 0.1% Tween 20 in Tris-buffered saline overnight at 4°C. Immunoblotting with anti-vimentin (Cell Signaling) was performed according to the manufacturer’s instructions (Amersham Biosciences). Signals were detected using a Storm 860 PhosphorImager system (Amersham Biosciences).
Two-dimensional gel electrophoresis (2-DE), in-gel tryptic digestion, and MALDI-TOF mass spectrometric analysis of RNA binding proteins
Purified proteins were resolved by 2-DE. 2-DE was performed as described by Görg et al., with minor modifications.Citation29 IPG strips were used according to the manufacturer's instruction. Isoelectric focusing (IEF) as the first dimension was performed on Protean IEF cell (Bio-Rad). Briefly, purified samples were mixed with an aliquot (185 μl) of rehydration solution [7 M urea, 2 M thiourea, 4% CHAPS (w/v), 60 mM DTT, a trace of bromophenol blue and 0.5% IPG buffer (v/v); Amersham Pharmacia Biotech] and then applied to IPG strips. After rehydration (12 h), IEF was performed with the following voltage-time program: 500 V for 1 h; 1000 V for 1 h: 8000 V over a gradient up to 50,000 V/h. Prior to SDS-PAGE, the IPG strips were incubated for 15 min with a solution of TRIS-HCl buffer (50 mM, pH 8.8), urea (6 M), glycerol (30%, v/v), SDS (2%, w/v), DTT (2%, w/v). Strips were then equilibrated for another 15 min in the same buffer that contained iodoacetamide (2.5%, w/v) instead of DTT. SDS-PAGE as the second dimension was performed at 90 V for 3 h. Molecular masses were determined by running standard protein markers (DualColor PrecisionPlus Protein™ standard; Bio-Rad). Duplicate 2-DE gels (control and sample) were run under the same conditions. Two gels were subjected to colloidal Coomassie staining (Simply Blue, Invitrogen) to visualize the protein spots, and gel slices of interest (differential bands) were subjected to in-gel tryptic digestion as described previously.Citation30 Tryptic peptides were extracted with 5% acetic acid followed by 5% acetic acid and 50% acetonitrile. Samples were dissolved in 5% acetic acid and desalted using ZipTip™ C18 reverse-phase desalting Eppendorf tips (Millipore). The peptides were eluted with 2% acetonitrile containing 0.1% TFA in a volume of 20 µl. Samples were analyzed using a MALDI-TOF mass spectrometer (Applied Biosystems). Tandem mass spectra were extracted and charge states deconvoluted by BioWorks version 2.0. Deisotoping was not performed. All MS/MS samples were analyzed using Sequest (Thermo Fisher Scientific; version 27, rev. 13) and X! Tandem (The GPM, thegpm.org; version 2007.01.01.1). X! Tandem was set up to search the rs_mus_v200810_cRAP database (35180 entries) assuming the digestion enzyme trypsin. Sequest was set up to search the rs_mus_v200810_cRAP database (35,180 entries) also assuming trypsin. Sequest and X! Tandem were searched with a fragment ion mass tolerance of 1.00 Da and a parent ion tolerance of 0.80 Da. The iodoacetamide derivative of cysteine was specified in Sequest and X! Tandem as a fixed modification. Oxidations of methionine and the iodoacetic acid derivative of cysteine were specified in Sequest and X! Tandem as variable modifications.
Criteria for protein identification
Scaffold (version Scaffold_3.6.0, Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm.Citation31 Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm.Citation32 Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
RNA electromobility shift analysis (REMSA)
REMSAs were performed as described,Citation33 with modifications. We used oligonucleotides corresponding to the wild type RNA sequences [Non B-175–150: 5′-UCAGACGUUCCAUUCUGCCUGCCGC-3′]. Briefly, the non-labeled RNPs (purified by RNA affinity purification) were incubated with [γ-32P]-labeled RNA oligonucleotides ( and ) in a final volume of 20 μl of REMSA buffer [10% glycerol, 1 mM MgCl2, 0.5 mM EDTA, 10 mM TRIS-HCl (pH 7.8), 0.1 mg/ml BSA and 0.5 mg/ml yeast tRNA], at room temperature for 30 min. Following formation of ribonucleoprotein (RNP) complexes, the REMSA reactions were electrophoresed on a 4% polyacrylamide gel in 0.5X TBE (45 mM Tris-borate and 1 mM EDTA) at 4°C and visualized by autoradiography.
For competition assays, the cell mixture was preincubated with 200 pmol (100X) of an unlabeled RNA for 5 min prior to the addition of the radiolabeled RNA. For antibody supershift assays, the reaction mixture was preincubated with 1 μg of the indicated antibody on ice for 10 min prior to the addition of the radiolabeled RNA. Purified vimentin protein was obtained from Abcam (Cambridge, MA, USA).
Heterogeneous expression of vimentin
For heterogeneous expression, the 4A-Vim plasmids were transfected into NS20Y and Neuro2A cells using the Effectene transfection reagent (Qiagen). To examine the regulation of MOR by vimentin, total proteins were isolated from the both cells transfected with vimentin. To correct for the differences in transfection efficiency, a one-fifth molar ratio of a pCH110 plasmid (Amersham) containing the β-galactosidase gene under the SV40 promoter was included in each transfection for normalization. The MOR (luciferase) and galactosidase (internal control) activities of each lysate were determined according to the manufacturer’s instructions (Promega and Tropix, respectively). The relative MOR mRNA (LUC) level was reported as the ratio of LUC mRNA/LacZ mRNA by real-time PCR.
Statistical analysis
At least three experiments were conducted to obtain statistical results, presented as means ± SD. Data were analyzed using the Student’s t-test, where p < 0.05 was considered to be signisficant.
Acknowledgments
This work was supported by the National Institutes of Health (Grants DA000564, DA001583, DA011806, DA011190 and DA023905) and by the A&F Stark Fund of the Minnesota Medical Foundation. The authors declare no conñict of interest.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Min BH, Augustin LB, Felsheim RF, Fuchs JA, Loh HH. Genomic structure analysis of promoter sequence of a mouse mu opioid receptor gene. Proc Natl Acad Sci U S A 1994; 91:9081 - 5; http://dx.doi.org/10.1073/pnas.91.19.9081; PMID: 8090773
- Wei LN, Loh HH. Regulation of opioid receptor expression. Curr Opin Pharmacol 2002; 2:69 - 75; http://dx.doi.org/10.1016/S1471-4892(01)00123-0; PMID: 11786311
- Law PY, Loh HH, Wei LN. Insights into the receptor transcription and signaling: implications in opioid tolerance and dependence. Neuropharmacology 2004; 47:Suppl 1 300 - 11; http://dx.doi.org/10.1016/j.neuropharm.2004.07.013; PMID: 15464146
- Wei LN, Law PY, Loh HH. Post-transcriptional regulation of opioid receptors in the nervous system. Front Biosci 2004; 9:1665 - 79; http://dx.doi.org/10.2741/1362; PMID: 14977578
- Kim CS, Hwang CK, Choi HS, Song KY, Law PY, Wei LN, et al. Neuron-restrictive silencer factor (NRSF) functions as a repressor in neuronal cells to regulate the mu opioid receptor gene. J Biol Chem 2004; 279:46464 - 73; http://dx.doi.org/10.1074/jbc.M403633200; PMID: 15322094
- Kim CS, Choi HS, Hwang CK, Song KY, Lee BK, Law PY, et al. Evidence of the neuron-restrictive silencer factor (NRSF) interaction with Sp3 and its synergic repression to the mu opioid receptor (MOR) gene. Nucleic Acids Res 2006; 34:6392 - 403; http://dx.doi.org/10.1093/nar/gkl724; PMID: 17130167
- Choi HS, Kim CS, Hwang CK, Song KY, Law PY, Wei LN, et al. Novel function of the poly(C)-binding protein alpha CP3 as a transcriptional repressor of the mu opioid receptor gene. FASEB J 2007; 21:3963 - 73; http://dx.doi.org/10.1096/fj.07-8561com; PMID: 17625070
- Song KY, Hwang CK, Kim CS, Choi HS, Law PY, Wei LN, et al. Translational repression of mouse mu opioid receptor expression via leaky scanning. Nucleic Acids Res 2007; 35:1501 - 13; http://dx.doi.org/10.1093/nar/gkm034; PMID: 17284463
- Hwang CK, Song KY, Kim CS, Choi HS, Guo XH, Law PY, et al. Evidence of endogenous mu opioid receptor regulation by epigenetic control of the promoters. Mol Cell Biol 2007; 27:4720 - 36; http://dx.doi.org/10.1128/MCB.00073-07; PMID: 17452465
- Choi HS, Hwang CK, Kim CS, Song KY, Law PY, Loh HH, et al. Transcriptional regulation of mouse mu opioid receptor gene in neuronal cells by poly(ADP-ribose) polymerase-1. J Cell Mol Med 2008; 12:6A 2319 - 33; http://dx.doi.org/10.1111/j.1582-4934.2008.00259.x; PMID: 18266974
- Choi HS, Song KY, Hwang CK, Kim CS, Law PY, Wei LN, et al. A proteomics approach for identification of single strand DNA-binding proteins involved in transcriptional regulation of mouse mu opioid receptor gene. Mol Cell Proteomics 2008; 7:1517 - 29; http://dx.doi.org/10.1074/mcp.M800052-MCP200; PMID: 18453338
- Song KY, Kim CS, Hwang CK, Choi HS, Law PY, Wei LN, et al. uAUG-mediated translational initiations are responsible for human mu opioid receptor gene expression. J Cell Mol Med 2010; 14:1113 - 24; PMID: 19438807
- Hwang CKSK, Song KY, Kim CS, Choi HS, Guo XH, Law PY, et al. Epigenetic programming of mu-opioid receptor gene in mouse brain is regulated by MeCP2 and Brg1 chromatin remodelling factor. J Cell Mol Med 2009; 13:9B 3591 - 615; http://dx.doi.org/10.1111/j.1582-4934.2008.00535.x; PMID: 19602036
- Song KY, Choi HS, Hwang CK, Kim CS, Law PY, Wei LN, et al. Differential use of an in-frame translation initiation codon regulates human mu opioid receptor (OPRM1). Cell Mol Life Sci 2009; 66:2933 - 42; http://dx.doi.org/10.1007/s00018-009-0082-7; PMID: 19609488
- Takada H, Kawana T, Ito Y, Kikuno RF, Mamada H, Araki T, et al. The RNA-binding protein Mex3b has a fine-tuning system for mRNA regulation in early Xenopus development. Development 2009; 136:2413 - 22; http://dx.doi.org/10.1242/dev.029165; PMID: 19542354
- Keene JD. Minireview: global regulation and dynamics of ribonucleic Acid. Endocrinology 2010; 151:1391 - 7; http://dx.doi.org/10.1210/en.2009-1250; PMID: 20332203
- Ray D, Kazan H, Chan ET, Peña Castillo L, Chaudhry S, Talukder S, et al. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat Biotechnol 2009; 27:667 - 70; http://dx.doi.org/10.1038/nbt.1550; PMID: 19561594
- Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci 2011; 68:3033 - 46; http://dx.doi.org/10.1007/s00018-011-0735-1; PMID: 21637948
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 2008; 582:1977 - 86; http://dx.doi.org/10.1016/j.febslet.2008.03.004; PMID: 18342629
- Mittal N, Roy N, Babu MM, Janga SC. Dissecting the expression dynamics of RNA-binding proteins in posttranscriptional regulatory networks. Proc Natl Acad Sci U S A 2009; 106:20300 - 5; http://dx.doi.org/10.1073/pnas.0906940106; PMID: 19918083
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature 2002; 416:499 - 506; http://dx.doi.org/10.1038/416499a; PMID: 11932736
- Keene JD, Tenenbaum SA. Eukaryotic mRNPs may represent posttranscriptional operons. Mol Cell 2002; 9:1161 - 7; http://dx.doi.org/10.1016/S1097-2765(02)00559-2; PMID: 12086614
- Keene JD. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc Natl Acad Sci U S A 2001; 98:7018 - 24; http://dx.doi.org/10.1073/pnas.111145598; PMID: 11416181
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science 2005; 309:1514 - 8; http://dx.doi.org/10.1126/science.1111443; PMID: 16141059
- Hogg JR, Collins K. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA 2007; 13:868 - 80; http://dx.doi.org/10.1261/rna.565207; PMID: 17456562
- Ermakova S, Choi BY, Choi HS, Kang BS, Bode AM, Dong Z. The intermediate filament protein vimentin is a new target for epigallocatechin gallate. J Biol Chem 2005; 280:16882 - 90; http://dx.doi.org/10.1074/jbc.M414185200; PMID: 15713670
- Suzuki T, Nakamoto T, Ogawa S, Seo S, Matsumura T, Tachibana K, et al. MICAL, a novel CasL interacting molecule, associates with vimentin. J Biol Chem 2002; 277:14933 - 41; PMID: 11827972
- Majumder M, Yaman I, Gaccioli F, Zeenko VV, Wang C, Caprara MG, et al. The hnRNA-binding proteins hnRNP L and PTB are required for efficient translation of the Cat-1 arginine/lysine transporter mRNA during amino acid starvation. Mol Cell Biol 2009; 29:2899 - 912; http://dx.doi.org/10.1128/MCB.01774-08; PMID: 19273590
- Görg A, Obermaier C, Boguth G, Harder A, Scheibe B, Wildgruber R, et al. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 2000; 21:1037 - 53; http://dx.doi.org/10.1002/(SICI)1522-2683(20000401)21:6<1037::AID-ELPS1037>3.0.CO;2-V; PMID: 10786879
- Patterson SD, Aebersold R. Mass spectrometric approaches for the identification of gel-separated proteins. Electrophoresis 1995; 16:1791 - 814; http://dx.doi.org/10.1002/elps.11501601299; PMID: 8586048
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 2002; 74:5383 - 92; http://dx.doi.org/10.1021/ac025747h; PMID: 12403597
- Nesvizhskii AIKA, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 2003; 75:4646 - 58; http://dx.doi.org/10.1021/ac0341261; PMID: 14632076
- Song KY, Choi HS, Law PY, Wei LN, Loh HH. Post-transcriptional regulation of mu-opioid receptor: role of the RNA-binding proteins heterogeneous nuclear ribonucleoprotein H1 and F. Cell Mol Life Sci 2012; 69:599 - 610; http://dx.doi.org/10.1007/s00018-011-0761-z; PMID: 21739230