Abstract
The archaeal RNA-degrading exosome contains a catalytically active hexameric core, an RNA-binding cap formed by Rrp4 and Csl4 and the protein annotated as DnaG (bacterial type primase) with so-far-unknown functions in RNA metabolism. We found that the archaeal DnaG binds to the Csl4-exosome but not to the Rrp4-exosome of Sulfolobus solfataricus. In vitro assays revealed that DnaG is a poly(A)-binding protein enhancing the degradation of adenine-rich transcripts by the Csl4-exosome. DnaG is the second poly(A)-binding protein besides Rrp4 in the heteromeric, RNA-binding cap of the S. solfataricus exosome. This apparently reflects the need for effective and selective recruitment of adenine-rich RNAs to the exosome in the RNA metabolism of S. solfataricus.
Keywords: :
Introduction
The RNA processing and degrading protein complex named exosome was originally described in eukarya where it is essential.Citation1 The existence of its archaeal counterpart was predicted based on the presence of genes encoding orthologs of the subunits of the eukaryotic exosome Rrp4, Rrp41, Rrp42 and Csl4 in most archaeal genomes.Citation2 Direct purification of the exosome from the thermoacidophilic archaeon Sulfolobus solfataricus confirmed the presence of a protein complex containing the Rrp4, Rrp41, Rrp42 and Csl4 orthologs. Surprisingly, the archaeal DnaG protein was co-purified with the S. solfataricus exosome, and later on it was shown that DnaG is an integral part of this protein complex.Citation3,Citation4 DnaG was also co-purified with the exosomes from two further archaeal species, Methanothermobacter thermoautotrophicus and Thermococcus kodakarensis, which belong to different phylogenetic lines.Citation5,Citation6 The annotation of the archaeal DnaG protein as a bacterial type primase is based on its central TOPRIM domain, which is characteristic for topoisomerases and primases.Citation7 Recently it was published that DnaG of S. solfataricus exhibits primase activity in vitro.Citation8 Since S. solfataricus also harbors a eukaryotic-type primase consisting of two subunits, it was proposed that this organism uses a dual system of primases.Citation8-Citation10 However, the strong association of DnaG with the exosome suggests that this protein plays an important role in RNA metabolism in the third domain of life.
The structure of the archaeal exosome without DnaG is well-understood. The subunits Rrp41 and Rrp42 form a phosphorolytically active hexamer, on the top of which a trimeric, RNA-binding cap is located.Citation11-Citation15 The crystallized archaeal exosomes have homomeric caps of either Rrp4 or Csl4. Rrp4 contains an N-terminal domain, a central S1-domain and a C-terminal KH-domain, while Csl4 contains an N-terminal domain, a central S1-domain and a C-terminal Zn-ribbon domain.Citation2 So far, crystallization of the archaeal exosome with heteromeric, Rrp4 and Csl4 containing caps, was not achieved. The existence of exosomes with such heteromeric caps was shown in vitro for Achaeoglobus fulgidus and in vivo for S. solfataricus.Citation12,Citation16 However, the stoichiometry of the cap proteins in vivo is not clear. A major part of the S. solfataricus exosome is detectable at the cell periphery and in the insoluble fraction, and the exosomes in the soluble and the insoluble fractions contain different relative amounts of Rrp4, Csl4 and DnaG.Citation16,Citation17
Although the mechanism of the catalytic Rrp41-Rrp42 hexamer is well-understood, less is known about the specific roles of Rrp4 and Csl4. Crystal structure analyses revealed that an RNA substrate interacts with the RNA-binding cap and its 3′-end is threaded through the S1-pore into the central channel of the hexamer, where it reaches one of the three active sites.Citation13-Citation15 In line with this, the additional recruitment of Rrp4 or Csl4 to the S. solfataricus hexamer results in increased RNA degradation in vitro.Citation4 However, the two cap proteins differently influence poly(A)-degradation by reconstituted exosomes of different archaeal species.Citation4,Citation12,Citation18-Citation20 It can be expected that the presence of two different proteins in the RNA-binding cap of the exosome enables interactions with different substrates. Indeed, it was shown recently that Rrp4 and Csl4 confer different specificities to the S. solfataricus exosome: The complex with a homomeric Rrp4 cap (the Rrp4-exosome) strongly prefers poly(A)-RNA, while the Csl4-exosome does not show this preference in RNA degradation assays.Citation21
The strong poly(A)-specificity of the Rrp4-exosome was surprising, since S. solfataricus lacks homopolymeric poly(A)-tails at the 3′-end of RNA.Citation22 Generally, post-transcriptionally synthesized poly(A)-tails and adenine-rich tails are important determinants of RNA stability in eukaryotic and prokaryotic cells.Citation23,Citation24 Archaea do not harbor homomeric poly(A)-tails at all, but exosome-containing Archaea have heteropolymeric, adenine-rich tails, which are believed to be synthesized by the exosome and to destabilize RNA.Citation22 It is assumed that in this respect the archaeal exosome functionally resembles prokaryotic polynucleotide phosphorylase (PNPase), to which it also shows high structural similarity.Citation11,Citation12 Prokaryotic PNPase is a phosphorolytic 3′-5′ exoribonuclease, which, in a reverse reaction, uses NDPs to add adenine-rich, destabilizing tails to the 3′-end of RNAs.Citation24,Citation25 The destabilizing function of an adenine-rich tail of a 16S rRNA fragment from S. solfataricus was demonstrated in vitro, but the effect was dependent on the composition of the RNA-binding cap of the exosome: The RNA-tail enhanced the degradation of a heteropolymeric transcript by the Rrp4-exosome but not by the Csl4-exosome, showing that the poly(A)-specificity of Rrp4 enables an efficient interaction with adenine-rich substrates.Citation21,Citation22
In addition to their different functions in recruitment of substrates, Rrp4 and Csl4 may differently interact with other proteins that co-purify with the exosome.Citation4-Citation6,Citation16 Recently it was shown in vitro that the U- and AU-binding Nip7p homolog from Pyrococcus abyssi binds better to the Csl4-exosome than to the Rrp4-exosome and negatively influences the activity of the complex.Citation19 Despite the strong association of DnaG with the exosome, the mechanisms and the physiological relevance of this interaction have not been explored so far.Citation4,Citation16 The aim of this work was to analyze the binding of DnaG to the archaeal exosome in vitro and to investigate the consequences of this interaction for RNA degradation.
Results
DnaG interacts with the Csl4-exosome
To study the mechanisms of interaction between DnaG and the exosome, we reconstituted complexes with homomeric RNA-binding caps containing either Rrp4 or Csl4 and tested them for interaction with DnaG by co-immunoprecipitation with Rrp41-specific antibodies. His-tagged versions of all proteins were used for these tests. shows that the Rrp4-exosome does not interact with DnaG, and demonstrates a clear binding of DnaG to the Csl4-exosome.
Figure 1. DnaG needs Csl4 for the interaction with the exosome. Silver-stained 12% SDS-gels showing results of experiments analyzing the interaction between DnaG and the exosome. In, input, the mixture of proteins used; FT, flow-through; W1, W5, the first and the last washing fractions; E, the elution fraction. (A) Rrp4-exosome or Csl4-exosome was mixed with DnaG and the interacting proteins were co-immunoprecipitated with Rrp41-specific antibodies. His-tagged proteins were used. (B) The Rrp41-Rrp42 hexamer (hexameric ring) alone or mixed with DnaG was subjected to co-immunoprecipitation (CoIP) with DnaG-specific antibodies. His-tagged proteins were used. The protein fractions were analyzed by SDS-PAGE and silver staining (upper panel) and by western blot hybridization with Rrp41-specific antibodies. (C) Strep-tagged Csl4 and His-tagged DnaG were mixed and a pull-down assay with Strep-Tactin Sepharose beads was performed. M, protein marker, the migration behavior of the proteins is given in kDa on the left side. An E. coli protein binding to Ni-NTA, which was routinely present in the DnaG-fractions used for the reconstitution experiment, is marked with an asterisk in (A and C). (D) His-tagged Rrp4 and His-tagged DnaG were mixed and CoIP with Rrp4-specific antibodies was performed. The detected proteins are marked on the right side. (E) Reconstituted and purified Csl4-exosome and DnaG-Csl4-exosome. (F) Reconstituted and purified Rrp4-Csl4-exosome and DnaG-Rrp4-Csl4-exosome. For the experiments shown in (E and F), Strep-Csl4 and His-tagged Rrp4, Rrp41, Rrp42 and DnaG proteins were used. Purification was performed with Strep-Tactin followed by Ni-NTA chromatography.
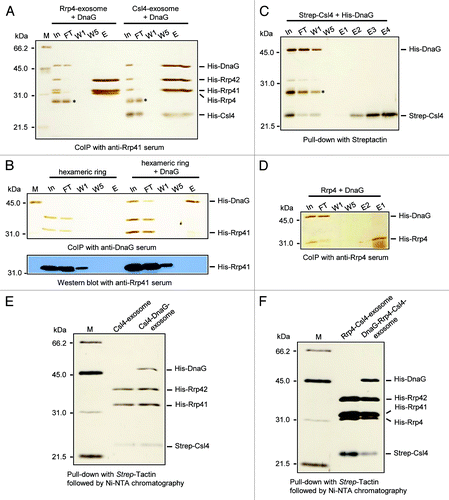
Next, we tested whether DnaG interacts with the Rrp41-Rrp42 hexamer using DnaG-specific antibodies (upper panel in ). In the elution fraction of this co-immunoprecipitation experiment (lane E at the right of the panel) we observed as expected DnaG, and a weak band migrating like Rrp41. However, this band was not Rrp41, as revealed by western blot analysis of the protein fractions (lower panel in ). The sensitivity of the Rrp41-specific antibodies was sufficient, since Rrp41 was detected in the first washing fraction W1 in the western blot analysis, although it was below the limit of detection by silver staining (compare the upper and the lower panels in ). We conclude that DnaG does not interact with the hexameric ring. To test whether DnaG interacts with Csl4, a strep-tagged version of Csl4 was mixed with the His6-DnaG, and pull-down assays with Strep-Tactin beads were performed (). Unexpectedly, no interaction between Csl4 and DnaG was detected. In line with the results shown in , DnaG also did not interact with Rrp4 ().
Since Rrp4 and Csl4 form heteromeric RNA-binding caps in vivo, we decided to test whether DnaG binds to reconstituted exosomal complexes with such heteromeric caps.Citation16 The strep-tagged Csl4 was mixed with the other His-tagged subunits of the exosome and Strep-Tactin Sepharose beads were used to fish Strep-Csl4 together with interacting proteins. Then Ni-NTA chromatography was applied to remove unbound Strep-Csl4. shows the successfully reconstituted and purified Strep-Csl4-exosome with and without DnaG. Thus, the interaction of DnaG with the Csl4-exosome was demonstrated by an independent method, in addition to the co-immunoprecipitation experiment shown in . When His6-Rrp4 and Strep-Csl4 were used together in reconstitution experiments, both proteins were co-purified by the tandem chromatography, demonstrating the formation of S. solfataricus exosomes with heteromeric caps in vitro. Furthermore, DnaG was found to form a stable complex in vitro with the exosome containing heteromeric caps (). This result is in line with the finding that DnaG interacts with the Rrp4-Csl4-exsosome in vivo.Citation16 Together with it confirms that Csl4 is necessary for the interaction of DnaG with the archaeal exosome.
DnaG modulates the substrate specificity of the Csl4-exosome
To study the function of DnaG in RNA metabolism, we compared the RNA-degrading properties of the Csl4-exosome in the presence and in the absence of DnaG using a 30-meric poly(A)-RNA as substrate. The reaction mixtures were incubated at 60°C in the presence of inorganic phosphate (Pi) and resolved on 16% polyacrylamide gels (). The exosome degraded the substrate to final products with a length of 4–5 nt.Citation26 Intermediates with lengths between 15 and 6 nt were most probably products of a distributive degradation in contrast to the processive degradation of longer molecules.Citation20,Citation26 The percentage of the remaining 30-meric poly(A) RNA per lane was determined. The data show that DnaG stimulates the degradation of the substrate by the Csl4-exosome (). As a negative control, the experiments were performed with the Rrp4-exosome, which does not interact with DnaG. In agreement with previous results, a 25 nt intermediate degradation product was detected in the assays with the Rrp4-exosome ().Citation21 Quantification of the remaining 30-meric poly(A)-RNA revealed that DnaG did not influence its degradation by the Rrp4-exosome (). Thus, the positive effect of DnaG on the degradation of the 30-meric poly(A)-RNA by the Csl4-exosome is specific.
Figure 2. DnaG influences the degradation properties of the Csl4-exosome but not of the Rrp4-exosome, and confers poly(A) specificity to the Csl4-exosome. (A) A phosphorimage of a 16% polyacrylamide gel with degradation assays containing 8 pmol radioactively labeled poly(A) 30-mer and 0.3 pmol of the Csl4-exosome or the Csl4-exosome supplemented with DnaG. The time of incubation at 60°C is also indicated (in min). The poly(A) substrate and the degradation products (degr. products) are marked on the right side. The size of the degradation products was previously estimated.Citation26 Control, negative control without protein. (B) Graphical representation of the results shown in (A) and from two additional independent experiments. (C) A phosphorimage of a 16% polyacrylamide gel with degradation assays containing 8 pmol radioactively labeled poly(A) 30-mer and 0.3 pmol of the Rrp4-exosome or the Rrp4-exosome supplemented with DnaG. For further descriptions, see (A). (D) Graphical representation of the results shown in (C) and from an additional independent experiment. (E‒G) Graphs showing the relative amount of the remaining substrate (in %) against the time (in min) in degradation assays (data from three independent experiments). In each reaction, 0.3 pmol protein complex was used. The protein complexes, labeled RNAs and non-labeled competitors, and their amounts per reaction mixture are indicated. (H) A phosphorimage of a 16% polyacrylamide gel with degradation assays containing the proteins indicated above the panel. 0.3 pmol of either Rrp41, Rrp42, Rrp4, Csl4, DnaG, the hexameric ring or the DnaG-Csl4-exosome were incubated for 10 min at 60°C with 8 fmol (lanes 1‒7) or 1 pmol (lanes 8‒12) radioactively labeled poly(A) 30-mer. Control, negative control without protein. Only His-tagged proteins were used for the experiments in this figure.
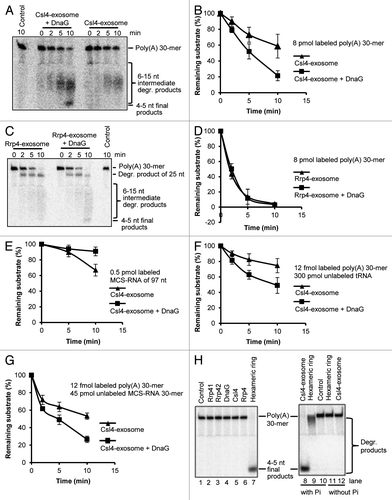
In the next experiment, a heteropolymeric in vitro transcript of 97 nt named MCS-RNA was used to compare the degradation properties of the Csl4-exosome in the presence and in the absence of DnaG.Citation26 The adenine content of MCS-RNA is relatively low (23%), and its predicted 3′-single stranded region of 16 nt contains only three adenines.Citation26 Previously, it was shown that the Csl4-exosome does not prefer poly(A)-RNA over MCS-RNA.Citation21 We found that DnaG negatively influences the degradation of this substrate by the Csl4-exosome ().
Competition experiments with labeled poly(A)-RNA and an excess of non-labeled, heteropolymeric RNA confirmed the higher efficiency of poly(A)-degradation by the DnaG-Csl4-exosome. As competitors, we used unlabeled yeast tRNA () and an MCS-RNA derivative of 30 nt corresponding to the 3′-part of full-length MCS-RNA (). The tRNAs are longer than the labeled poly(A)-RNA and are highly structured, while the 30-meric MCS-RNA derivative has the same length but is predicted to form only weak secondary structures. It was shown previously that both competitors can be degraded by the Csl4-exosome.Citation21,Citation26
The observed RNase activity was not due to contamination of the recombinant S. solfataricus proteins by bacterial RNases, since the preparations of the hexameric ring, Rrp4, Csl4 and DnaG were heat treated at 75°C for 20 min prior to usage. Even the non-heat treated preparations of Rrp41 and Rrp42 did not show RNase activities under the applied reaction temperature of 60°C (, lanes 2 and 3). We have shown previously that at this temperature, RNases in the cell-free extract of E. coli are easily inactivated.Citation26 In agreement with previous data, the individual subunits of the exosome could not degrade RNA (, lanes 2‒6), and the assembly of the Rrp41/Rrp42 hexamer was necessary for activity (lane 7). As expected, the degradation efficiency of the Csl4-exosome was higher than the activity of the hexamer (compare lane 8 to lane 9), and inorganic phosphate was necessary for RNA degradation (lanes 8‒12).Citation4,Citation11 In conclusion, the controls in show that the phosphorolytic degradation of RNA in our assays is due to the hexameric core of the archaeal exosome.
In summary, shows that DnaG modulates the substrate specificity of the Csl4-exosome: The DnaG-Csl4-exosome degrades MCS-RNA with lower efficiency than the Csl4-exosome, while poly(A) RNA is degraded with higher efficiency. We conclude that DnaG confers poly(A) specificity to the Csl4-exosome.
DnaG is a poly(A)-binding protein
The results shown in raised the question whether DnaG is a poly(A)-binding subunit of the S. solfataricus exosome. This question was addressed by electrophoretic mobility shift assays (EMSA) aiming to compare the poly(A)-binding properties of the Csl4-exosome and of DnaG. shows that the 30-meric poly(A)-RNA (25 fmol per reaction) was bound much stronger by DnaG than by the Csl4-exosome when comparable molar amounts of the proteins were used. For example, the substrate was shifted as a stable band by 1.2 pmol DnaG, while no shift was observed with the same amount of Csl4-exosome (compare lanes 2 and 9 in ). To analyze the poly(A)-specificity of DnaG, competition experiments were performed (). Addition of 11 pmol or 22 pmol of unlabeled, 30-meric MCS RNA did not abolish the binding of 25 fmol labeled poly(A)-RNA by 2.5 pmol of DnaG (compare lane 1 to lanes 3 and 5). In contrast, when unlabeled poly(A)-RNA was added to the reaction mixtures, the labeled poly(A)-RNA was detected only in its unbound form (lanes 7 and 9). These results show that DnaG preferentially binds poly(A)-RNA.
Figure 3. Poly(A)-RNA binding by DnaG, the Csl4-exosome and the DnaG-Csl4-exosome. Shown are phosphorimages of EMSA assays in native 5% polyacrylamide gels. Twenty-five fmol labeled poly(A)-RNA 30-mer was used in each reaction. Only His-tagged proteins were used for the experiments in (A and B). Strep-tagged Csl4 was used for the reconstitution of the complexes (see ) used in (C and D). (A) Binding assays with the Csl4-exosome or with DnaG in different amounts as indicated above the panel. His-tagged proteins were used. (B) Binding of DnaG (amount indicated above each lane) to radiolabeled poly(A) 30-mer in the absence or presence of unlabeled competitor RNAs. Lanes 1, 2 and 11, no competitor was used. Lanes 3‒10, competitors in the following amounts were used: Lanes 3 and 4, 11 pmol MCS-RNA; lanes 5 and 6; 22 pmol MCS-RNA; lanes 7 and 8, 11 pmol poly(A); lanes 9 and 10, 22 pmol poly(A). (C) Binding assays with 2.5 pmol of DnaG, the Csl4-exosome or the DnaG-Csl4-exosome, as indicated above the panel. The amounts of the unlabeled MCS-RNA-derived 30-mer used as competitor are also indicated above the corresponding lanes. (D) Binding assays with 0.6 pmol of DnaG, the Csl4-exosome or the DnaG-Csl4-exosome, as indicated above the panel. The percentages of unbound poly(A) substrate in each lane is indicated below the panels (results from three independent experiments). The proportion of unbound substrate in the control lane was set to 100%. Control, negative control without protein and without competitor. The migration in the gel of the unbound poly(A) 30-mer and of the protein/RNA complexes (complexes) is indicated on the right side.
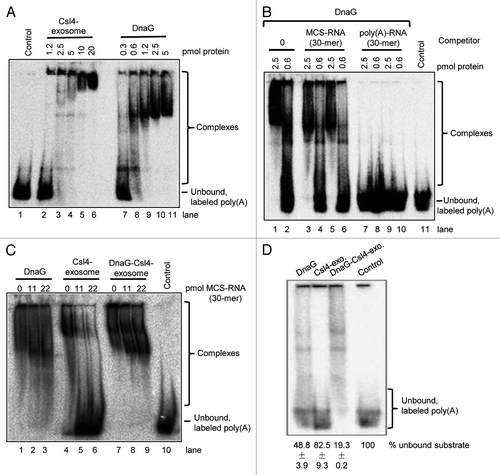
Next, poly(A)-binding assays in the presence and in the absence of unlabeled MCS, RNA were performed with DnaG, the Csl4-exosome and the DnaG-Csl4-exosome (with 2.5 pmol of DnaG and of each complex). The exosomal complexes used in these assays are shown in . The complex purification with Strep-Tactin beads followed by Ni-NTA chromatography ensured that no free DnaG was present in the preparation of the DnaG-Csl4-exosome. shows that when DnaG or the DnaG-Csl4-exosome were used, no free poly(A)-RNA was detected in the presence of the unlabeled MCS RNA competitor (see lanes 1‒3 and 7‒9). Thus, DnaG and the DnaG-Csl4-exosome show binding preference for poly(A). In contrast, the labeled poly(A)-RNA was not bound by the Csl4-exosome when unlabeled MCS RNA was added to the reaction mixture (lanes 5 and 6). This is consistent with the finding that the Csl4-exosome does not prefer poly(A)-RNA over MCS RNA.Citation21 We conclude that DnaG confers poly(A)-binding specificity to the Csl4-exosome.
To test whether the Csl4-exosome increases the binding activity of DnaG, similar binding assays were performed with lower protein amounts (0.6 pmol). The labeled poly(A) 30-mer was bound very weakly by the Csl4-exosome, 82.5 ± 9.3% remained unbound in comparison to the negative control set to 100% (). Consistent with the stronger binding of poly(A) by DnaG, approximately half of the substrate was shifted by DnaG and only 48.8 ± 3.9% remained unbound. Since more than 80% of the substrate was shifted by the DnaG-Csl4-complex (only 19.3 ± 0.2 remained unbound), we conclude the binding activity of DnaG is higher in the context of the exosome ().
Our results show that DnaG is a poly(A)-binding protein and a part of the RNA-binding platform of the archaeal exosome. Thus, the RNA-binding cap of the S. solfataricus exosome contains two poly(A)-binding proteins, Rrp4 and DnaG.Citation21
DnaG helps the Csl4-exosome to recruit naturally occurring, adenine-rich RNA
Since S. solfataricus does not harbor homopolymeric poly(A)-tails, we assumed that the function of DnaG in the RNA-binding cap of the S. solfataricus exosome is to ensure an efficient interaction with adenine-rich substrates.Citation22 To test this, we compared the degradation of MCS-RNA with the degradation of its tailed variant MCS-RNA19hetero by the Csl4-exosome and by the DnaG-Csl4-exosome. The latter substrate contains a heteromeric, adenine-rich tail of 19 nt (47% A-content of the tail), which was originally detected at the 3′-end of a 16S rRNA fragment in S. solfataricus.Citation21,Citation22 We found that the DnaG-Csl4-exosome degrades MCS-RNA19hetero with higher efficiency than MCS-RNA (). Conversely, the Csl4-exosome even tended to prefer the MCS-RNA as substrate (). In previous assays, the Csl4-exosome did not show clear preference for one of these substrates.Citation21 Thus, the DnaG-Csl4-exosome can better degrade the substrate with the adenine-rich tail than the non-tailed substrate, similarly to what was previously shown for the Rrp4-exosome.Citation21
Figure 4. DnaG enhances the interaction between heteropolymeric, adenine-rich transcripts and the Csl4-exosome. The relative amount of the remaining labeled substrate (in %) is plotted over time (in min). (A) Degradation assays with 0.3 pmol of the DnaG-Csl4-exosome, 0.2 pmol MCS-RNA (97 nt) and 0.2 pmol MCS-RNA carrying a heteropolymeric, adenine-rich tail of 19 nt (MCS-RNA19hetero). The two substrates were present together in reaction mixtures, in which one of the substrates was internally labeled and the other was unlabeled.Citation21 (B) Assays with 0.3 pmol of the Csl4-exosome and the substrates described in (A). (C) Assays with the DnaG-Csl4-exosome (0.3 pmol) and the Csl4-exosome (0.3 pmol) using 0.5 pmol labeled A-rich transcript (59 nt). (D) Assays with the DnaG-Csl4-exosome (0.3 pmol) and the Csl4-exosome (0.3 pmol) using 0.2 pmol labeled A-rich transcript (59 nt) and 0.2 pmol unlabeled MCS-RNA (97 nt). Graphs represent results from three independent experiments in (A and B), and two independent experiments in (C and D). His-tagged proteins were used for the experiments in this figure.
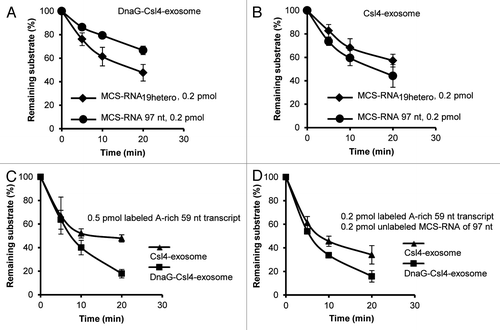
We also synthesized another in vitro transcript of 59 nt, which corresponds to a tail found at the 3′-end of a 16S rRNA fragment in S. solfataricus, and tested it in degradation assays.Citation22 In agreement with the poly(A) specificity of DnaG, this adenine-rich transcript (A-content of 64%) was degraded with better efficiency by the DnaG-Csl4-exosome than by the Csl4-exosome (). Similar results were obtained when MCS-RNA was added as competitor to the reaction mixture, confirming the preference of the DnaG-containing complex for adenine-rich RNA (). shows that DnaG increases the efficiency of interaction between the Csl4-exosome and adenine-rich RNAs.
DnaG increases the efficiency of the Rrp4-Csl4-exosome on poly(A) RNA
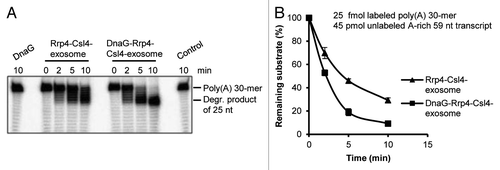
In vivo Csl4 and Rrp4 are forming together the RNA-binding cap of the soluble exosome.Citation16 Thus, the question arose whether DnaG can influence the interaction of the Rrp4-Csl4-exosome with poly(A)-RNA, since this exosome already contains the poly(A)-binding protein Rrp4.Citation21 This was tested in degradation assays with the Rrp4-Csl4-exosome and the DnaG-Rrp4-Csl4-exosome (). The used protein complexes are shown in . A substrate mixture of 25 fmol labeled, 30-meric poly(A)-RNA and 45 pmol unlabeled, adenine-rich transcript of 59 nt was used. shows that the poly(A)-RNA was converted into a 25 nt degradation intermediate routinely observed in degradation assays with the Rrp4-exosome (compare with ). This intermediate accumulates when longer substrates are present in the reaction mixture.Citation21 The degradation of the 30-meric poly(A)-RNA was faster in the presence of DnaG (). We conclude that DnaG enhances the efficiency of interaction between poly(A)-RNA and the exosome containing Rrp4 and Csl4.
Figure 5. DnaG enhances the interaction between poly(A)-RNA and the Rrp4-Csl4-exosome. (A) A phosphorimage of a 16% polyacrylamide gel with degradation assays containing 25 fmol radioactively labeled poly(A) 30-mer and 45 pmol unlabeled A-rich transcript (59 nt). 0.3 pmol of DnaG, the Rrp4-Csl4-exosome or the DnaG-Rrp4-Csl4-exosome were present in each reaction mixture as indicated above the panel. The Strep-Csl4-containing exosomes shown in were used. The incubation time (in min) at 60°C is indicated. The 30-meric poly(A) substrate and the accumulating degradation product of 25 nt (see ref. Citation21) are marked on the right side. Control, negative control without protein. (B) Graphical representation of the results shown in (A) and from two additional independent experiments.
Discussion
The analysis of the interaction between DnaG and the exosome of S. solfataricus revealed that DnaG binds only to Csl4- or Csl4-Rrp4-exosomes, while no binding to Csl4 alone, the Rrp41-Rrp42 hexamer or the Rrp4-exosome was detected (). A comparison between the crystal structures of the Csl4-exosome and the Rrp4 exosome from A. fulgidus did not reveal differences on the surface of the hexameric ring, suggesting that conformational changes of the hexamer upon Csl4 binding are not responsible for the specific interaction of DnaG with the Csl4-exosome.Citation12 Binding of DnaG to composite surfaces generated by complexation of Csl4 and the hexamer may explain why DnaG binds to the Csl4-exosome but not to Csl4 or the hexamer. Alternatively or in addition, Csl4 may change its conformation upon binding to the Rrp41-Rrp42 hexamer, allowing a strong interaction with DnaG. Similarly, monomeric S. solfataricus Rrp4 does not interact with a short poly(A)-RNA of 7 nt, while this poly(A)-RNA is bound 40-fold stronger by the Rrp4-exosome than by the Rrp41-Rrp42 hexamer.Citation27 Taken together, the data suggest that both Csl4 and Rrp4 are inactive in their monomeric states, but adopt their active conformations in the context of the archaeal exosome.
Our finding that DnaG interacts with reconstituted exosomes containing heteromeric RNA-binding caps composed of Rrp4 and Csl4 agrees with the detection of exosomal complexes containing all three proteins, Rrp4, Csl4 and DnaG, in the soluble fraction of S. solfataricus.Citation16 The fact that the interaction of DnaG with the exosome depends on Csl4 is in line with the finding that increased proportions of DnaG and Csl4 are present in the insoluble form of the exosome, suggesting a functional relationship between the two proteins.Citation16
In addition to the uncovering of the interaction partner of DnaG in the exosome, we elucidated the function of DnaG in the protein complex. Our data show that DnaG is a part of the RNA-binding cap of the archaeal exosome and influences its interaction with RNA substrates. The degradation of most of the labeled substrates analyzed here was increased when DnaG was added to the Csl4-exosome. We can exclude that binding of DnaG to the Csl4-exosome simply stabilizes the protein complex leading to better RNA degradation, since the adenine-poor MCS-RNA was degraded with lower efficiency by the DnaG-Csl4-exosome than by the Csl4-exosome (). These results show that DnaG modulates the substrate specificity of the Csl4-exosome leading to the preference for adenine-rich substrates.
The identification of S. solfataricus DnaG as a poly(A)-RNA-binding protein was unexpected. The presence of two poly(A)-binding proteins, Rrp4 and DnaG, in the RNA-binding cap of the S. solfataricus exosome suggests an evolutionary pressure for the presence of poly(A)-binding domains in this protein complex. The three-dimensional structure of the archaeal exosome resembles prokaryotic PNPase, which contains three S1-domains with poly(A)-binding capacity.Citation11,Citation12,Citation14,Citation28 The S1 domains in the archaeal nine-subunit exosome are provided by Rrp4 and Csl4, but only Rrp4 shows poly(A) specificity.Citation2,Citation16,Citation21 It can be assumed that many of the substrates of the exosome in S. solfataricus are adenine-rich mRNAs and adenine-rich, post-transcriptionally synthesized RNA-tails.Citation22,Citation29 Short poly(A) stretches in these substrates are likely the main recognition epitopes for Rrp4 and DnaG. Apparently, the RNA metabolism of S. solfataricus demands effective and selective degradation of such A-rich RNAs, explaining the evolutionary recruitment of another exosomal subunit in addition to Rrp4, which is able to capture such RNA substrates in order to thread them into the catalytic core of the archaeal exosome. This notion is in line with our finding that DnaG enhances the degradation of poly(A) RNA by the physiologically relevant Rrp4-Csl4-exosome ().
In conclusion, we show that Csl4 is necessary for the binding of the archaeal DnaG-protein to the exosome and assign a function to DnaG in the context of this protein complex. DnaG is the second poly(A)-binding subunit in the RNA-recruiting cap of the S. solfataricus exosome besides Rrp4, and it helps the exosome to interact with adenine-rich substrates, which are common in this organism.
Materials and Methods
Protein purification and protein-protein interaction studies
His-tagged Rrp41, Rrp42, Rrp4 and Csl4 of S. solfataricus were expressed in E. coli BL21 (DE3). His-tagged DnaG was expressed in E. coli ArcticExpressTM (DE3) (Agilent cat# 230192). Purification by Ni-NTA affinity chromatography (Qiagen cat# 30210) under native conditions, complex reconstitution and co-immunoprecipitation with polyclonal antibodies were previously described.Citation4,Citation6,Citation16,Citation30 The His-tags of the proteins were not cleaved. Briefly, Rrp41 and Rrp42 were mixed (0.5 mg each) in a final volume of 1.5 ml. The mixture was incubated for 2 h at room temperature in the P0 buffer containing 10 mM Tris, pH 7, 5 mM MgCl2, 0.5 mM EDTA, 200 mM NaCl, 5% glycerol, 0.05% Tween 20 and 0.2 mM DTT. Thereafter, DnaG and/or Rrp4 or Csl4 was added (0.7 mg each), and 2 h incubation at room temperature was performed. After heat treatment at 75°C for 10 min and centrifugation for 20 min at 13,000 g, the supernatants were used for co-immunoprecipitation with 0.04 g protein A-Sepharose beads with covalently coupled rabbit sera specified in .Citation4,Citation16
Strep-tagged Csl4 of S. solfataricus was expressed in E. coli BL21 (DE3). The cells were lysed by sonication and the cell-free extract was heated at 75°C for 20 min. Strep-tag Csl4 was purified by Strep-Tactin® Sepharose® (iba cat# 2-1201-010) according to the instructions of the manufacturer. Strep-tagged Csl4 (0.5 mg) and His-tagged DnaG (0.7 mg) were mixed and incubated as described above. The mixture was loaded onto Strep-Tactin sepharose column (1 ml), washed with 6-column bed volumes and eluted six times (with 100 μl each time). For reconstitution of Strep-Csl4-exosomes, Strep-tagged Csl4 (0.5 mg), His6-Rrp41 (1 mg) and His6-Rrp42 (1 mg) were mixed in buffer P0. When appropriate, His6-DnaG (1 mg) and 0.5 mg His6-Rrp4 were added. The mixtures were heat treated at 75°C for 10 min, and purification with Strep-Tactin was performed as described above. After dialysis with buffer P0, the protein complexes were purified by Ni-NTA affinity chromatography. Thereby, unbound Strep-Csl4 was removed and the exosome was concentrated. Aliquots were stored at -80°C, and second thawing was avoided.
Protein fractions were analyzed by SDS-PAGE and silver staining. For western blot analyses, samples were resolved in 12% SDS-PAGE and transferred to Protran nitrocellulose membrane (whatman cat# 10485376). The membrane was blocked in 1 × TBS containing 5% non-fat milk powder for 1 h at 4°C. Signals were detected with primary anti- Rrp41 antibodies and secondary anti-rabbit IgG conjugated with peroxidase (Sigma cat# A0545) by developing with Lumi-Light Western Blotting Substrate (Roche cat# 12015200001).
RNA degradation assays
Degradation assays using the reconstituted exosomal complexes were performed at 60°C in a reaction buffer containing 20 mM HEPES (pH 7.9), 60 mM KCl, 8 mM MgCl2, 0.1 mM EDTA; 2 mM DTT and 10 mM K2HPO4 as described.Citation21,Citation26 The reconstituted hexameric ring was heat treated at 75°C for 20 min, purified through size exclusion chromatography, aliquoted and stored at -80°C. Aliquots were supplemented with the same molar amounts of His-tagged Rrp4, Csl4 and DnaG (heat treated at 75°C for 20 min) and the resulting protein complexes were used at the concentration of 0.03 pmol/µl in 10 µl degradation reactions.Citation21,Citation26 The reconstitution of Strep-Csl4-containing exosomes is described above. The aliquoted complexes were stored at -80°C and thawed only once. Separation of substrate and reaction products in high-resolution polyacrylamide gels under denaturing conditions was performed using Rothiphorese Gel 30 (Roth cat# 3029.1). The enzymes, the substrates and their amounts per reaction mixture are given in the legends to the figures. The generation of 5′-end-labeled 30-meric poly(A)-RNA, internally labeled MCS-RNA of 97 nt and its derivatives and unlabeled MCS-RNA of 97 nt and 30 nt was also described.Citation21,Citation26 The template oligonucleotide 5′-TTTTTTTTTTATCTTTTCGCGCTTTCTATTCCACTAATTTTTTGTTT-TTACTTTTTCCCTATAGTGAGTCGTATTA-3′ was annealed to the T7-promoter oligonucleotide and used for in vitro transcription of the internally labeled, adenine-rich transcript of 59 nt. The full sequences of the used in vitro transcripts are: MCS-RNA 97 nt, 5′-GGGAGACCGGCAGATCTGATATCATCGATGAATTCGAGCTCGGTACCCGGGG-ATCCTCTAGAGTCGACCTGCAGGCATGCAAGCTTCAGCTGCTCGA-3′; MCS-RNA19hetero (116 nt) 5′-GGGAGACCGGCAGAUCUGAUAUCAUCGAUGAAUUCGAGCUCGGUACCCG-GGGAUCCUCUAGAGUCGACCUGCAGGCAUGCAAGCUUCAGCUGCUCGAGGGAUAAUUAUAAAAGGAG-3′; MCS-RNA 30 nt, 5′-GGGUGCAGGCAUGCAAGCUUCAGCUGCUCGA -3′; A-rich 59 nt transcript, 5′-GGGAAAAAGUAAAAACAAAAAAUUAGUGGAAUAGAAAG-CGCGAAAAGAUAAAAAAAAAA-3′.
RNA binding assays
Binding assays were performed essentially as published by Büttner et al.Citation12 For the assays in , the reconstituted and gel filtration-purified hexameric ring was mixed with equimolar amount of His-tagged Csl4 and incubated on ice for 20 min prior to the assays. For the assays in , Strep-Csl4-containing exosomes were used, which were reconstituted as described above. The assay buffer contained 20 mM HEPES (pH 7.9), 60 mM KCl, 10 mM MgCl2, 10% glycerine, 2 mM DTT and 0.1 mM EDTA. The volume of the reaction mixtures was 10 µl. The amounts of proteins and RNA substrates are indicated in . The reaction mixtures were incubated at room temperature for 5 min and were then analyzed in 5% polyacrylamide gels at 200 V and 4°C. Visualization was performed by phosphorimaging.Citation26
Acknowledgments
We are grateful to Michael A. Trakselis for sending us the plasmid for overexpression of His-tagged DnaG. We thank Verena Roppelt for cloning the Strep-tag variant of Csl4.This work was supported by DFG, grant Kl563/27-1, and the China Scholarship Council.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-->5′ exoribonucleases. Cell 1997; 91:457 - 66; http://dx.doi.org/10.1016/S0092-8674(00)80432-8; PMID: 9390555
- Koonin EV, Wolf YI, Aravind L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res 2001; 11:240 - 52; http://dx.doi.org/10.1101/gr.162001; PMID: 11157787
- Evguenieva-Hackenberg E, Walter P, Hochleitner E, Lottspeich F, Klug G. An exosome-like complex in Sulfolobus solfataricus.. EMBO Rep 2003; 4:889 - 93; http://dx.doi.org/10.1038/sj.embor.embor929; PMID: 12947419
- Walter P, Klein F, Lorentzen E, Ilchmann A, Klug G, Evguenieva-Hackenberg E. Characterization of native and reconstituted exosome complexes from the hyperthermophilic archaeon Sulfolobus solfataricus.. Mol Microbiol 2006; 62:1076 - 89; http://dx.doi.org/10.1111/j.1365-2958.2006.05393.x; PMID: 17078816
- Farhoud MH, Wessels HJ, Steenbakkers PJ, Mattijssen S, Wevers RA, van Engelen BG, et al. Protein complexes in the archaeon Methanothermobacter thermautotrophicus analyzed by blue native/SDS-PAGE and mass spectrometry. Mol Cell Proteomics 2005; 4:1653 - 63; http://dx.doi.org/10.1074/mcp.M500171-MCP200; PMID: 16037073
- Li Z, Santangelo TJ, Cuboňová L, Reeve JN, Kelman Z. Affinity purification of an archaeal DNA replication protein network. MBio 2010; 1:e00221 - 10; http://dx.doi.org/10.1128/mBio.00221-10; PMID: 20978540
- Aravind L, Leipe DD, Koonin EV. Toprim--a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res 1998; 26:4205 - 13; http://dx.doi.org/10.1093/nar/26.18.4205; PMID: 9722641
- Zuo Z, Rodgers CJ, Mikheikin AL, Trakselis MA. Characterization of a functional DnaG-type primase in archaea: implications for a dual-primase system. J Mol Biol 2010; 397:664 - 76; http://dx.doi.org/10.1016/j.jmb.2010.01.057; PMID: 20122937
- De Falco M, Fusco A, De Felice M, Rossi M, Pisani FM. The DNA primase of Sulfolobus solfataricus is activated by substrates containing a thymine-rich bubble and has a 3′-terminal nucleotidyl-transferase activity. Nucleic Acids Res 2004; 32:5223 - 30; http://dx.doi.org/10.1093/nar/gkh865; PMID: 15459292
- Lao-Sirieix SH, Bell SD. The heterodimeric primase of the hyperthermophilic archaeon Sulfolobus solfataricus possesses DNA and RNA primase, polymerase and 3′-terminal nucleotidyl transferase activities. J Mol Biol 2004; 344:1251 - 63; http://dx.doi.org/10.1016/j.jmb.2004.10.018; PMID: 15561142
- Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat Struct Mol Biol 2005; 12:575 - 81; http://dx.doi.org/10.1038/nsmb952; PMID: 15951817
- Büttner K, Wenig K, Hopfner KP. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell 2005; 20:461 - 71; http://dx.doi.org/10.1016/j.molcel.2005.10.018; PMID: 16285927
- Lorentzen E, Conti E. Structural basis of 3′ end RNA recognition and exoribonucleolytic cleavage by an exosome RNase PH core. Mol Cell 2005; 20:473 - 81; http://dx.doi.org/10.1016/j.molcel.2005.10.020; PMID: 16285928
- Lorentzen E, Dziembowski A, Lindner D, Seraphin B, Conti E. RNA channelling by the archaeal exosome. EMBO Rep 2007; 8:470 - 6; http://dx.doi.org/10.1038/sj.embor.7400945; PMID: 17380186
- Navarro MV, Oliveira CC, Zanchin NI, Guimarães BG. Insights into the mechanism of progressive RNA degradation by the archaeal exosome. J Biol Chem 2008; 283:14120 - 31; http://dx.doi.org/10.1074/jbc.M801005200; PMID: 18353775
- Witharana C, Roppelt V, Lochnit G, Klug G, Evguenieva-Hackenberg E. Heterogeneous complexes of the RNA exosome in Sulfolobus solfataricus.. Biochimie 2012; 94:1578 - 87; http://dx.doi.org/10.1016/j.biochi.2012.03.026; PMID: 22503705
- Roppelt V, Hobel CF, Albers SV, Lassek C, Schwarz H, Klug G, et al. The archaeal exosome localizes to the membrane. FEBS Lett 2010; 584:2791 - 5; http://dx.doi.org/10.1016/j.febslet.2010.05.013; PMID: 20488181
- Ramos CR, Oliveira CL, Torriani IL, Oliveira CC. The Pyrococcus exosome complex: structural and functional characterization. J Biol Chem 2006; 281:6751 - 9; http://dx.doi.org/10.1074/jbc.M512495200; PMID: 16407194
- Luz JS, Ramos CR, Santos MC, Coltri PP, Palhano FL, Foguel D, et al. Identification of archaeal proteins that affect the exosome function in vitro. BMC Biochem 2010; 11:22; http://dx.doi.org/10.1186/1471-2091-11-22; PMID: 20507607
- Hartung S, Niederberger T, Hartung M, Tresch A, Hopfner KP. Quantitative analysis of processive RNA degradation by the archaeal RNA exosome. Nucleic Acids Res 2010; 38:5166 - 76; http://dx.doi.org/10.1093/nar/gkq238; PMID: 20392821
- Roppelt V, Klug G, Evguenieva-Hackenberg E. The evolutionarily conserved subunits Rrp4 and Csl4 confer different substrate specificities to the archaeal exosome. FEBS Lett 2010; 584:2931 - 6; http://dx.doi.org/10.1016/j.febslet.2010.05.014; PMID: 20488184
- Portnoy V, Evguenieva-Hackenberg E, Klein F, Walter P, Lorentzen E, Klug G, et al. RNA polyadenylation in Archaea: not observed in Haloferax while the exosome polynucleotidylates RNA in Sulfolobus.. EMBO Rep 2005; 6:1188 - 93; http://dx.doi.org/10.1038/sj.embor.7400571; PMID: 16282984
- Eckmann CR, Rammelt C, Wahle E. Control of poly(A) tail length. Wiley Interdiscip Rev RNA 2011; 2:348 - 61; http://dx.doi.org/10.1002/wrna.56; PMID: 21957022
- Slomovic S, Schuster G. Exonucleases and endonucleases involved in polyadenylation-assisted RNA decay. Wiley Interdiscip Rev RNA 2011; 2:106 - 23; http://dx.doi.org/10.1002/wrna.45; PMID: 21956972
- Mohanty BK, Kushner SR. Polynucleotide phosphorylase functions both as a 3′ right-arrow 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc Natl Acad Sci USA 2000; 97:11966 - 71; http://dx.doi.org/10.1073/pnas.220295997; PMID: 11035800
- Evguenieva-Hackenberg E, Roppelt V, Finsterseifer P, Klug G. Rrp4 and Csl4 are needed for efficient degradation but not for polyadenylation of synthetic and natural RNA by the archaeal exosome. Biochemistry 2008; 47:13158 - 68; http://dx.doi.org/10.1021/bi8012214; PMID: 19053279
- Oddone A, Lorentzen E, Basquin J, Gasch A, Rybin V, Conti E, et al. Structural and biochemical characterization of the yeast exosome component Rrp40. EMBO Rep 2007; 8:63 - 9; http://dx.doi.org/10.1038/sj.embor.7400856; PMID: 17159918
- Yehudai-Resheff S, Portnoy V, Yogev S, Adir N, Schuster G. Domain analysis of the chloroplast polynucleotide phosphorylase reveals discrete functions in RNA degradation, polyadenylation, and sequence homology with exosome proteins. Plant Cell 2003; 15:2003 - 19; http://dx.doi.org/10.1105/tpc.013326; PMID: 12953107
- De Vendittis E, Bocchini V. Protein-encoding genes in the sulfothermophilic archaea Sulfolobus and Pyrococcus. Gene 1996; 176:27 - 33; http://dx.doi.org/10.1016/0378-1119(96)00203-X; PMID: 8918227
- Lorentzen E, Conti E. Expression, reconstitution, and structure of an archaeal RNA degrading exosome. Methods Enzymol 2008; 447:417 - 35; http://dx.doi.org/10.1016/S0076-6879(08)02220-9; PMID: 19161854